| Original Literature | Model OverView |
|---|---|
|
Publication
Title
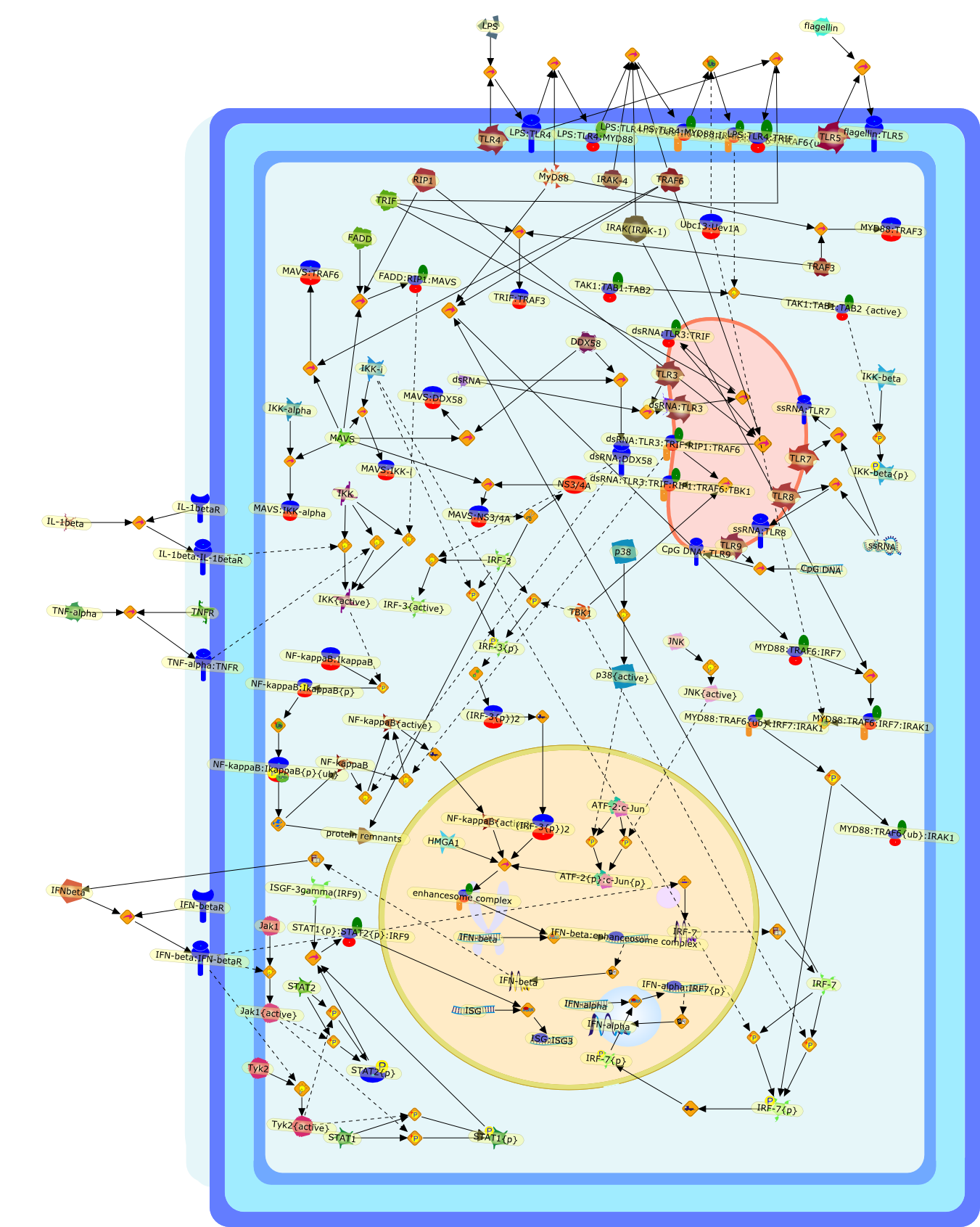
Antiviral innate immunity pathways.
Affiliation
Howard Hughes Medical Institute, Department of Molecular Biology, University ofTexas Southwestern Medical Center, Dallas, TX 75390-9148, USA.
Abstract
Recent studies have uncovered two signaling pathways that activate the hostinnate immunity against viral infection. One of the pathways utilizes members ofthe Toll-like receptor (TLR) family to detect viruses that enter the endosomethrough endocytosis. The TLR pathway induces interferon production throughseveral signaling proteins that ultimately lead to the activation of thetranscription factors NF-kappaB, IRF3 and IRF7. The other antiviral pathway usesthe RNA helicase RIG-I as the receptor for intracellular viral double-strandedRNA. RIG-I activates NF-kappaB and IRFs through the recently identified adaptorprotein MAVS, a CARD domain containing protein that resides in the mitochondrialmembrane. MAVS is essential for antiviral innate immunity, but it also serves asa target of Hepatitis C virus (HCV), which employs a viral protease to cleaveMAVS off the mitochondria, thereby allowing HCV to escape the host immunesystem.
PMID
16474426
|