| Original Literature | Model OverView |
|---|---|
|
Publication
Title
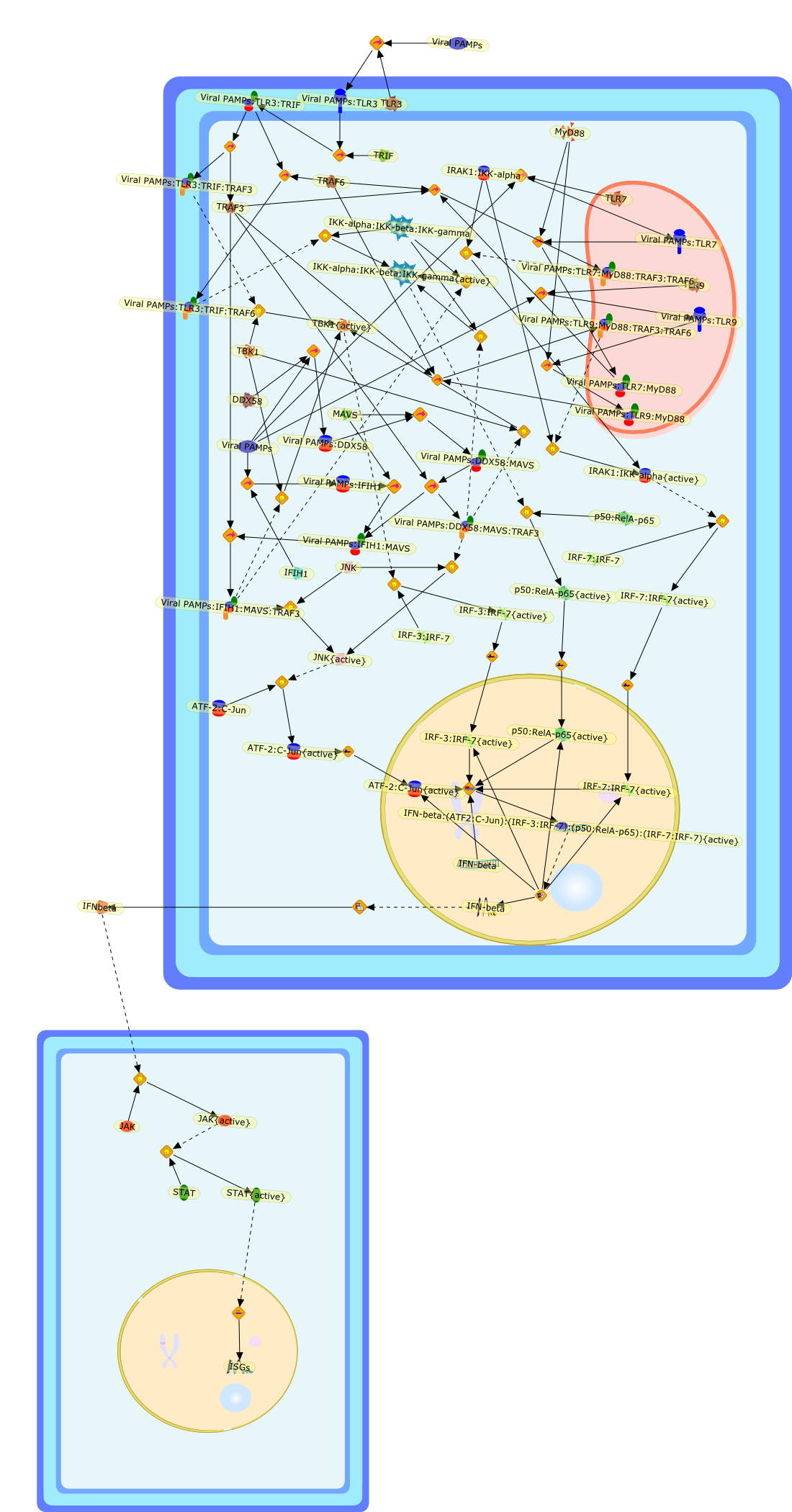
Parallel pathways of virus recognition.
Affiliation
Department of Molecular and Cellular Biology, Harvard University, Cambridge,Massachusetts 02138, USA.
Abstract
Viruses trigger signaling pathways of innate immunity. In this issue, it isshown that the mitochondrial antiviral signaling protein is critical forintracellular detection signaling, but is dispensable for the activation ofinnate immunity via Toll-like receptors.
PMID
16713969
|