| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Cellular reprogramming by gram-positive bacterial components: a review.
Affiliation
Department of Academic Surgery, Cork University Hospital, National University ofIreland/University College Cork, Wilton, Cork, Ireland.
Abstract
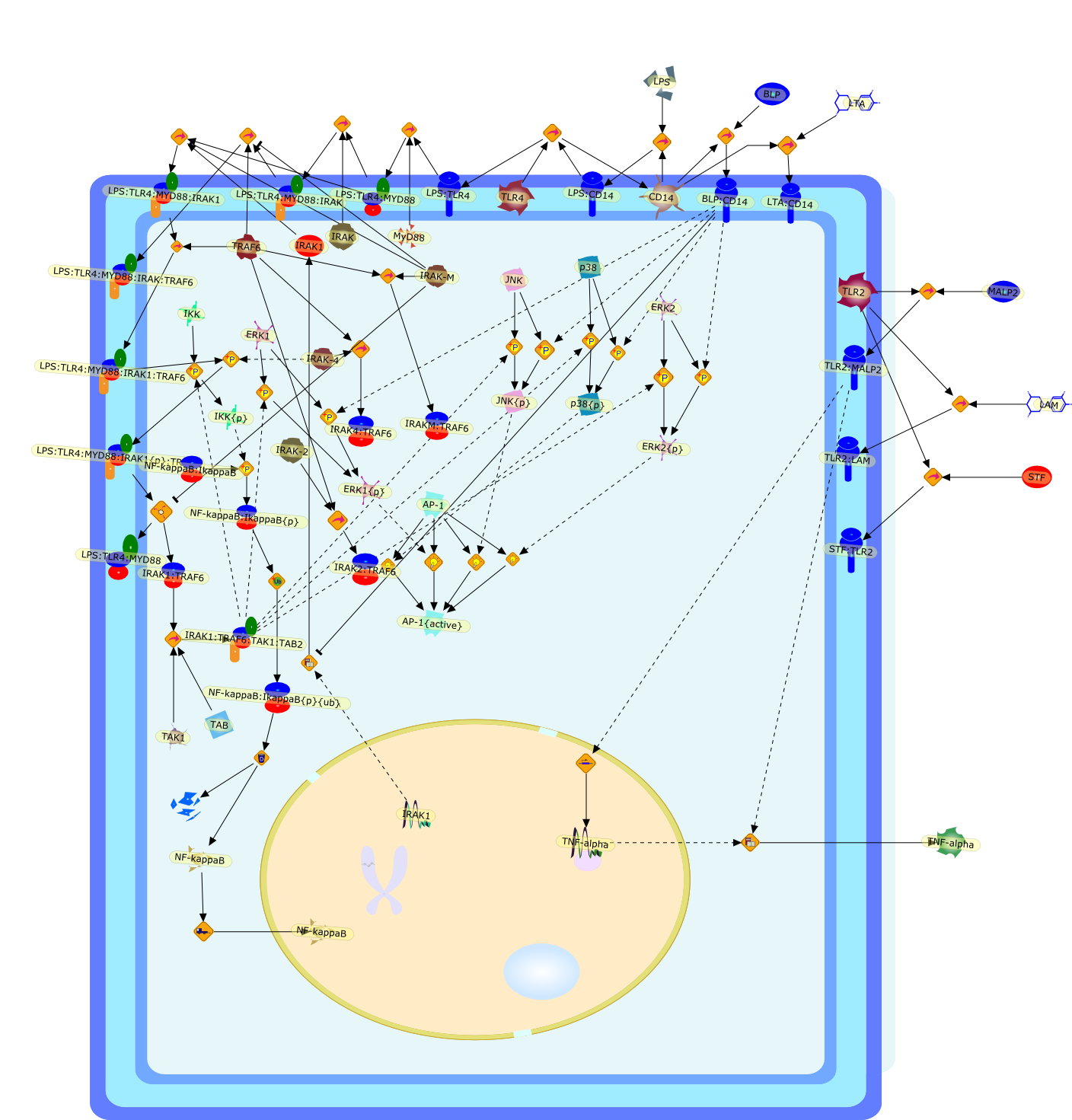
LPS tolerance has been the focus of extensive scientific and clinical researchover the last several decades in an attempt to elucidate the sequence of changesthat occur at a molecular level in tolerized cells. Tolerance to components ofgram-positive bacterial cell walls such as bacterial lipoprotein andlipoteichoic acid is a much lesser studied, although equally important,phenomenon. This review will focus on cellular reprogramming by gram-positivebacterial components and examines the alterations in cell surface receptorexpression, changes in intracellular signaling, gene expression and cytokineproduction, and the phenomenon of cross-tolerance.
PMID
16885502
|