| Original Literature | Model OverView |
|---|---|
|
Publication
Title
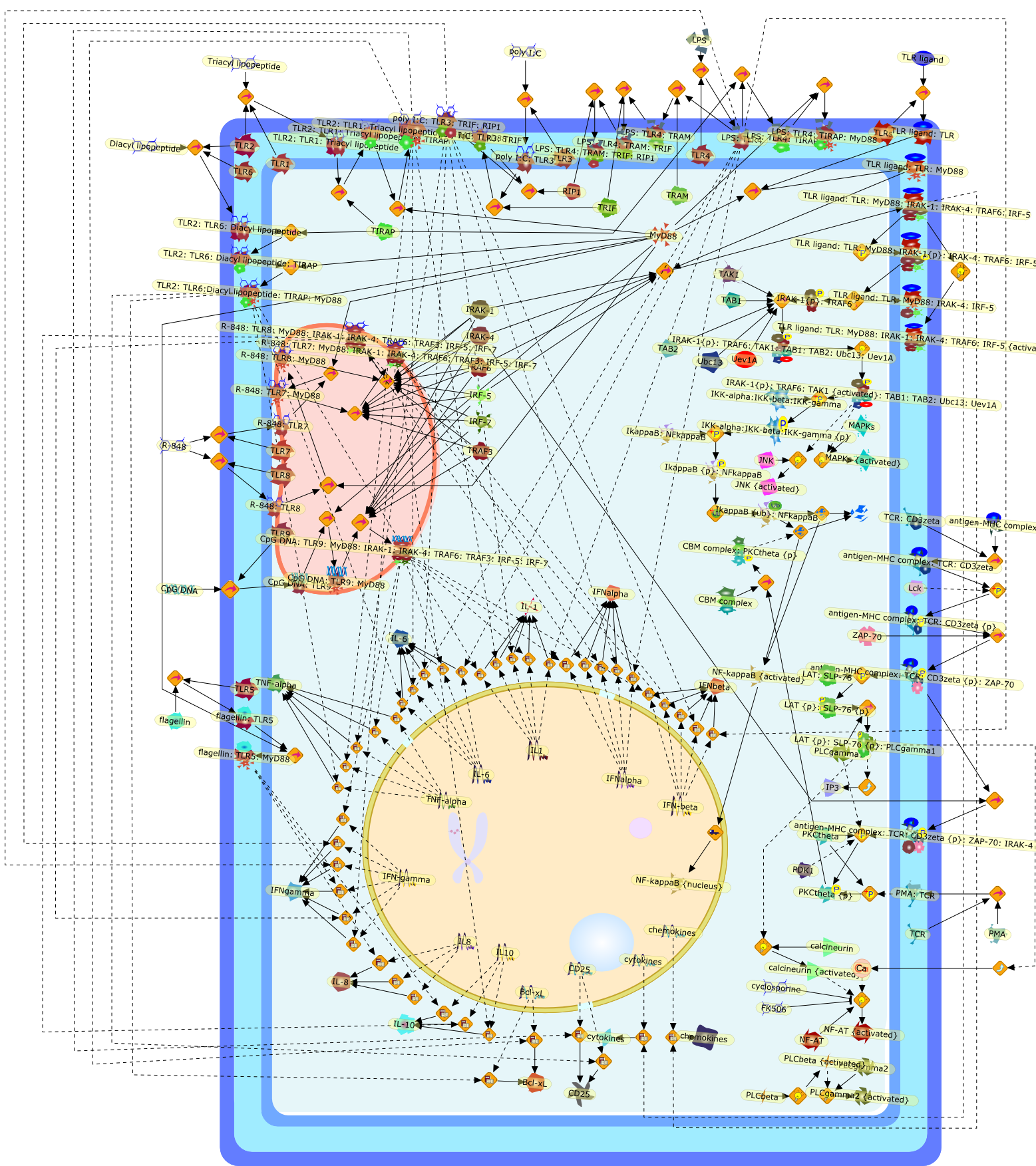
IRAK-4--a shared NF-kappaB activator in innate and acquired immunity.
Affiliation
Laboratory for Cell Signaling, RIKEN Research Center for Allergy and Immunology,1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan. nobu@rcai.riken.jp
Abstract
The human body is protected against external pathogens by two immune systems:innate and acquired immunities. Whereas innate immunity exhibits immediateresponses to external pathogens by recognizing pathogen-associated molecularpatterns (PAMPs), adaptive immunity uses T cells to recognize and defend againstpathogens by developing effector cells, antibodies and memory cells. Althougheach system seems to possess distinct activation mechanisms, interleukin-1receptor-associated kinase (IRAK)-4 is essential for NF-kappaB activation inToll-like receptor (TLR) and T-cell receptor (TCR) signaling pathways. Thisimplies possible crosstalk between innate and acquired immunities, andevolutionary development that resulted in the use of innate signaling moleculesby the acquired immune system. Here, we discuss the impact of theseevolutionarily conserved molecules on innate and acquired immunity, and theirpotential as drug targets for the simultaneous modulation of both immunities.
PMID
17046325
|