| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Shaping of monocyte and macrophage function by adenosine receptors.
Affiliation
Department of Surgery, UMDNJ-New Jersey Medical School, Newark, NJ 07103, USA.haskoge@umdnj.edu
Abstract
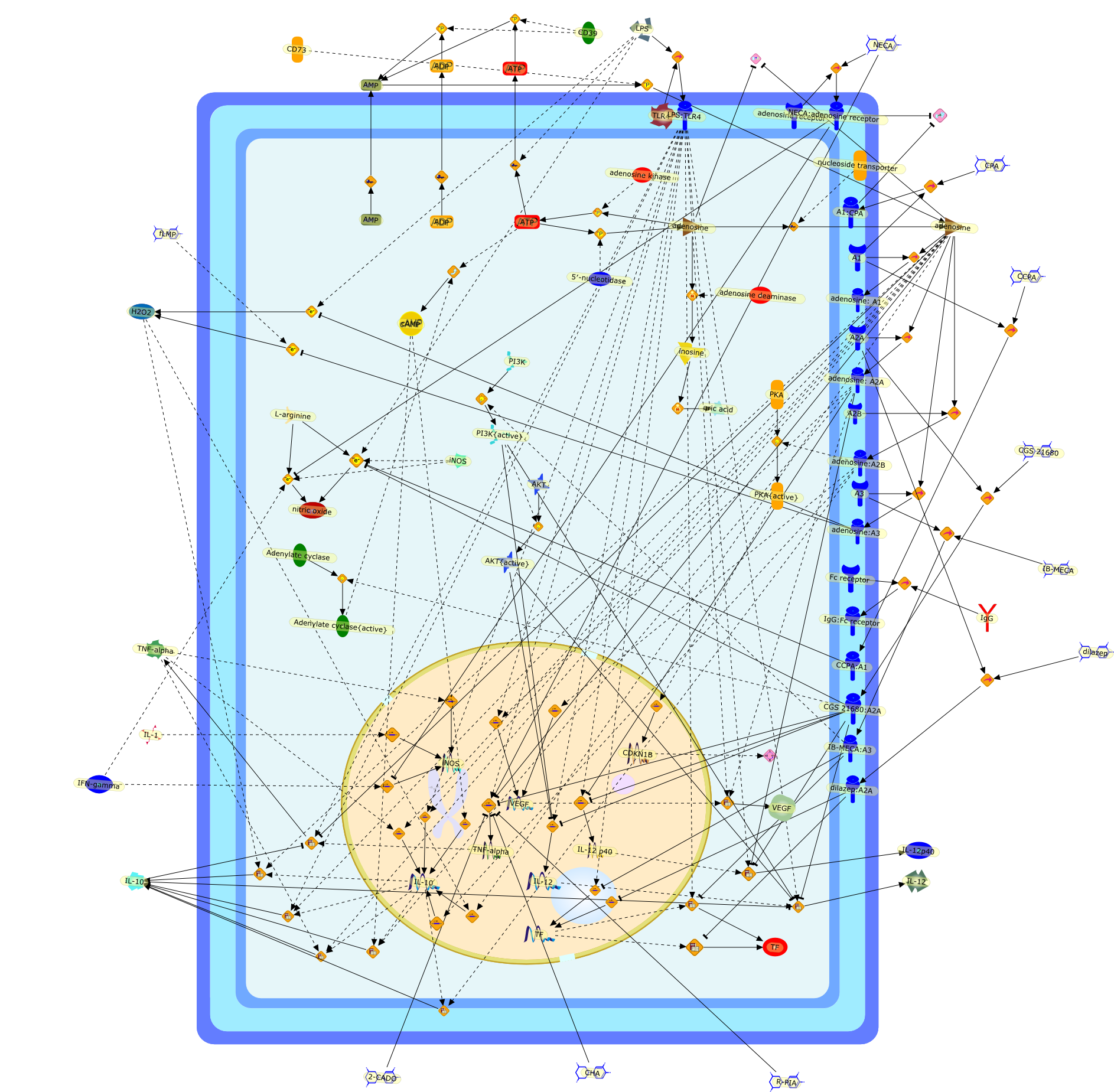
Adenosine is an endogenous purine nucleoside that, following its release intothe extracellular space, binds to specific adenosine receptors expressed on thecell surface. Adenosine appears in the extracellular space under metabolicallystressful conditions, which are associated with ischemia, inflammation, and celldamage. There are 4 types of adenosine receptors (A(1), A(2A), A(2B) and A(3))and all adenosine receptors are members of the G protein-coupled family ofreceptors. Adenosine receptors are expressed on monocytes and macrophages andthrough these receptors adenosine modulates monocyte and macrophage function.Since monocytes and macrophages are activated by the same danger signals thatcause accumulation of extracellular adenosine, adenosine receptors expressed onmacrophages represent a sensor system that provide monocytes and macrophageswith information about the stressful environment. Adenosine receptors, thus,allow monocytes and macrophages to fine-tune their responses to stressfulstimuli. Here, we review the consequences of adenosine receptor activation onmonocyte/macrophage function. We will detail the effect of stimulating thevarious adenosine receptor subtypes on macrophage differentiation/proliferation,phagocytosis, and tissue factor (TF) expression. We will also summarize ourknowledge of how adenosine impacts the production of extracellular mediatorssecreted by monocytes and macrophages in response to toll-like receptor (TLR)ligands and other inflammatory stimuli. Specifically, we will delineate howadenosine affects the production of superoxide, nitric oxide (NO), tumornecrosis factor-alpha, interleukin (IL)-12, IL-10, and vascular endothelialgrowth factor (VEGF). A deeper insight into the regulation of monocyte andmacrophage function by adenosine receptors should assist in developing newtherapies for inflammatory diseases.
PMID
17056121
|