| Original Literature | Model OverView |
|---|---|
|
Publication
Title
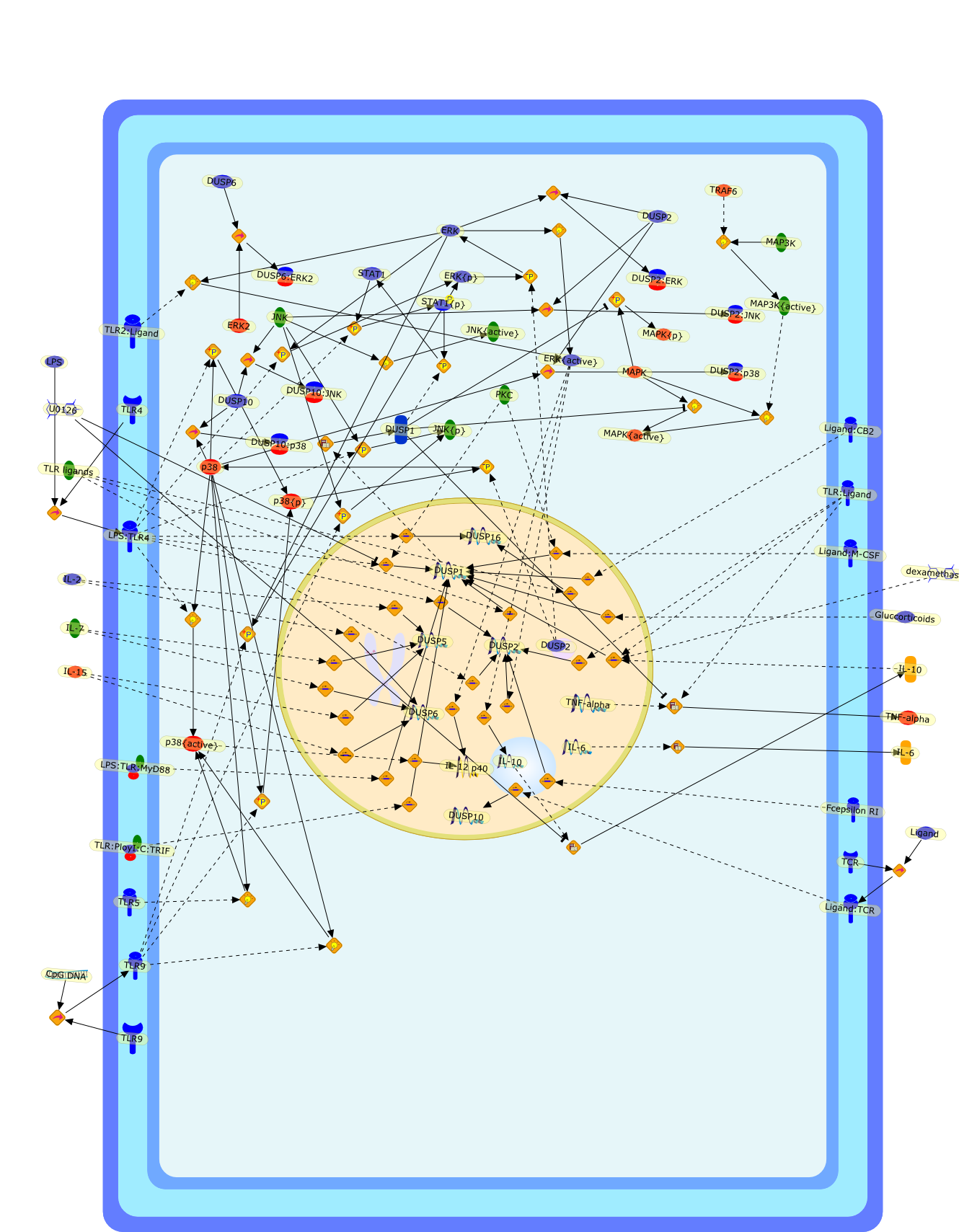
DUSP meet immunology: dual specificity MAPK phosphatases in control of theinflammatory response.
Affiliation
Institute of Medical Microbiology, Immunology and Hygiene, Technical UniversityMunich, Immunology and Hygiene, Trogerstrasse 30, Munich 81675, Germany.Roland.Lang@lrz.tum.de
Abstract
The MAPK family members p38, JNK, and ERK are all activated downstream of innateimmunity's TLR to induce the production of cytokines and inflammatory mediators.However, the relative intensity and duration of the activation of different MAPKappears to determine the type of immune response. The mammalian genome encodes alarge number of dual specificity phosphatases (DUSP), many of which act as MAPKphosphatases. In this study, we review the emergence of several DUSP as genesthat are differentially expressed and regulated in immune cells. Recently, aseries of investigations in mice deficient in DUSP1, DUSP2, or DUSP10 revealedspecificity in the regulation of the different MAPK proteins, and definedessential roles in models of local and systemic inflammation. The DUSP family isproposed as a set of molecular control devices specifying and modulating MAPKsignaling, which may be targeted to unleash or attenuate innate and adaptiveimmune effector functions.
PMID
17114416
|