| Original Literature | Model OverView |
|---|---|
|
Publication
Title
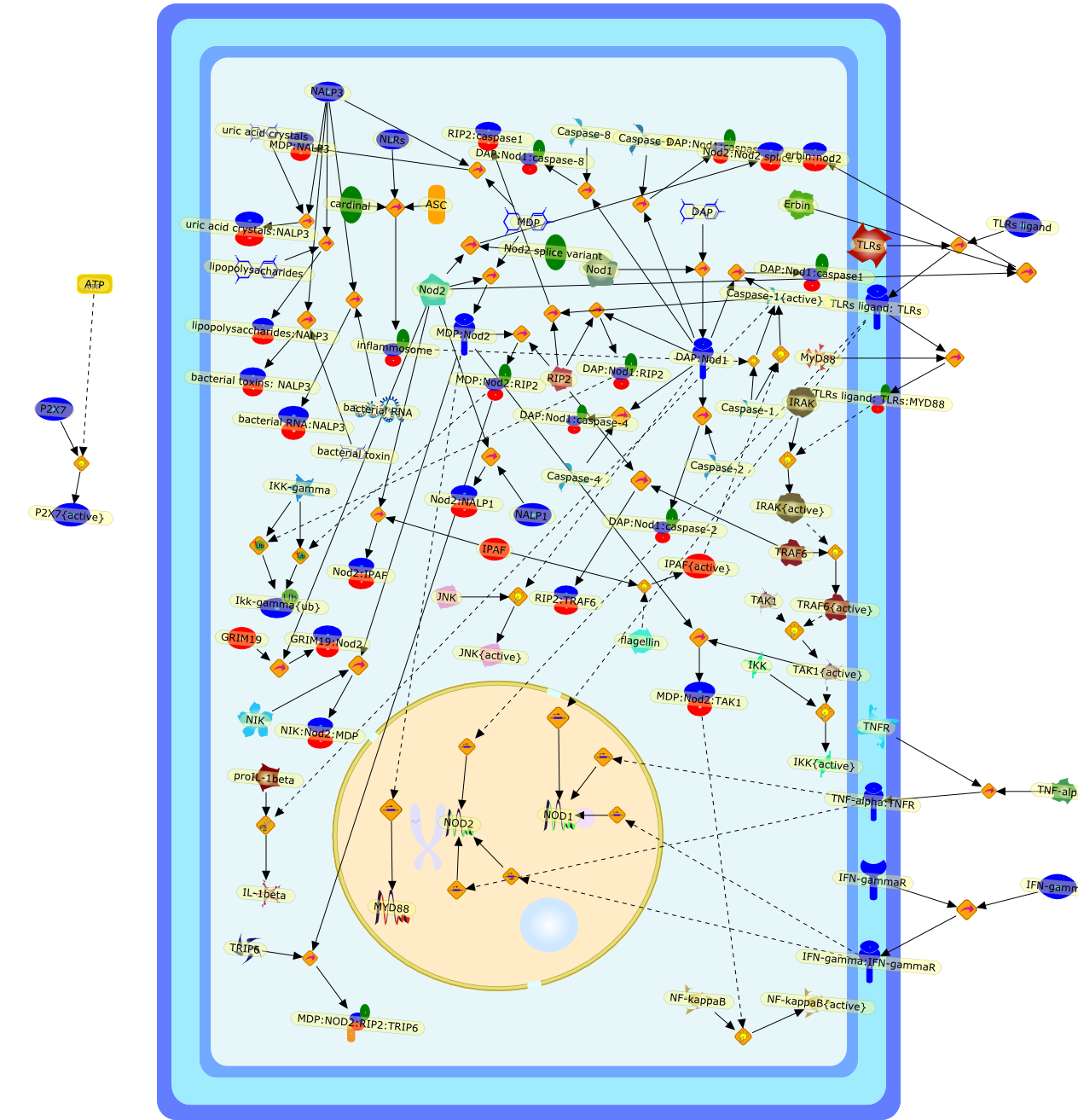
Sensing of bacteria: NOD a lonely job.
Affiliation
Unite de Pathogenie Microbienne Moleculaire, Institut Pasteur, F-75724 ParisCedex 15, France.
Abstract
Recognition of bacteria by the vertebrate innate immune system relies ondetection of invariant molecules by specialized receptors. The view is emergingthat activation of both Toll-like receptors (TLRs) and Nod-like receptors (NLRs)by different bacterial agonists is important in order to mount an inflammatoryresponse in the host. Priming of cells with peptidoglycan and products that aresensed by cytosolic-localized members of the NLR family have a synergisticeffect on TLR signalling and vice versa. Currently, the underlying molecularmechanisms of this cross-talk between NLR and TLR signalling are beginning toemerge. These reveal that the two sensing-systems are non-redundant in bacterialrecognition and that their cross-talk plays an important role in immunologicalhomeostasis.
PMID
17161646
|