| Original Literature | Model OverView |
|---|---|
|
Publication
Title
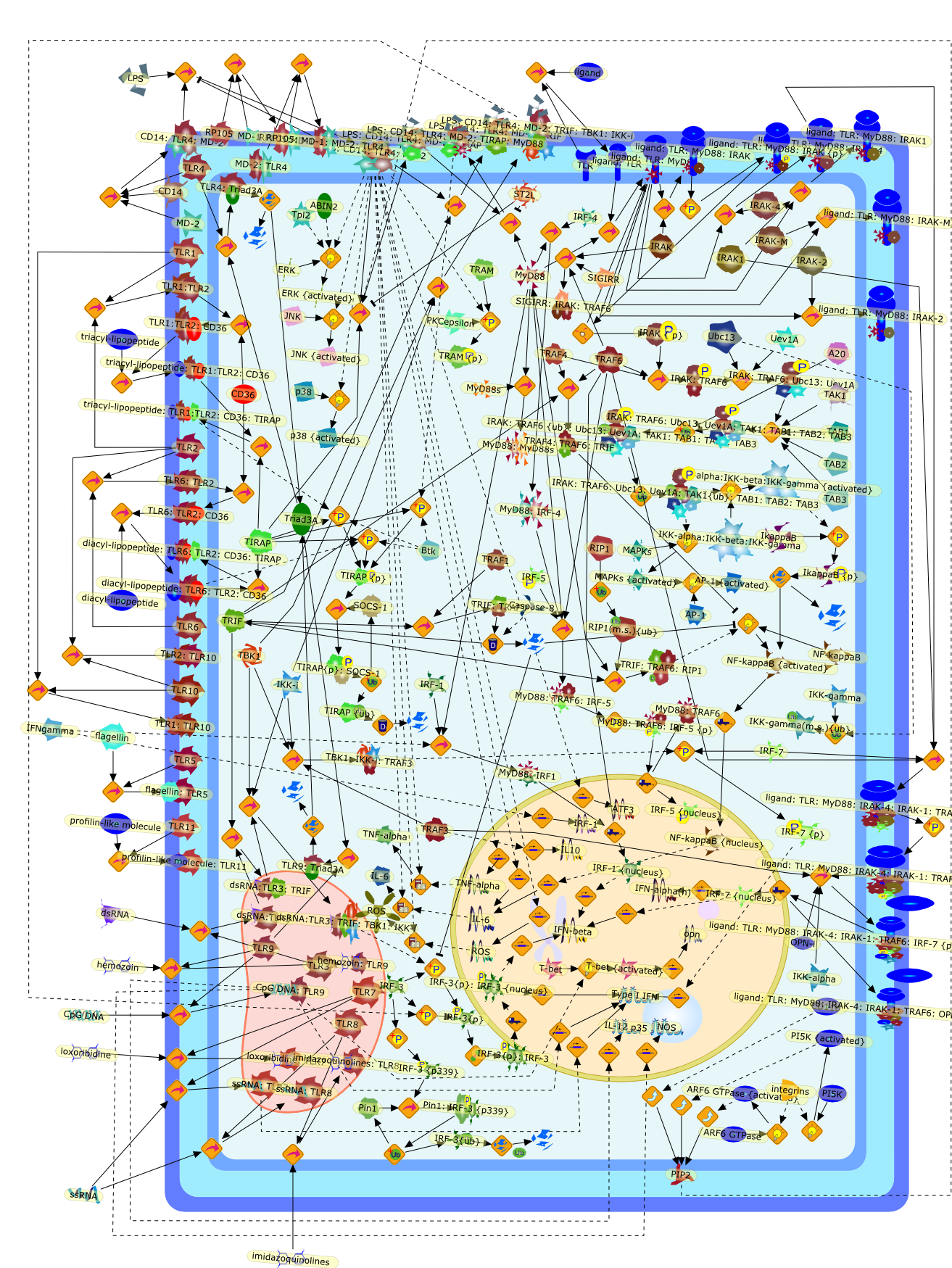
TLR signaling.
Affiliation
Department of Host Defense, Research Institute for Microbial Diseases, OsakaUniversity, Yamada-oka, Suita, Osaka, Japan.
Abstract
The TLR family senses the molecular signatures of microbial pathogens, and playsa fundamental role in innate immune responses. TLRs signal via a common pathwaythat leads to the expression of diverse inflammatory genes. In addition, eachTLR elicits specific cellular responses to pathogens owing to differential usageof intracellular adapter proteins. Recent studies have revealed the importanceof the subcellular localization of TLRs in pathogen recognition and signaling.TLR signaling pathways is negatively regulated by a number of cellular proteinsto attenuate inflammation. Here, we describe recent advances in ourunderstanding of the regulation of TLR-mediated signaling.
PMID
17275323
|