| Original Literature | Model OverView |
|---|---|
|
Publication
Title
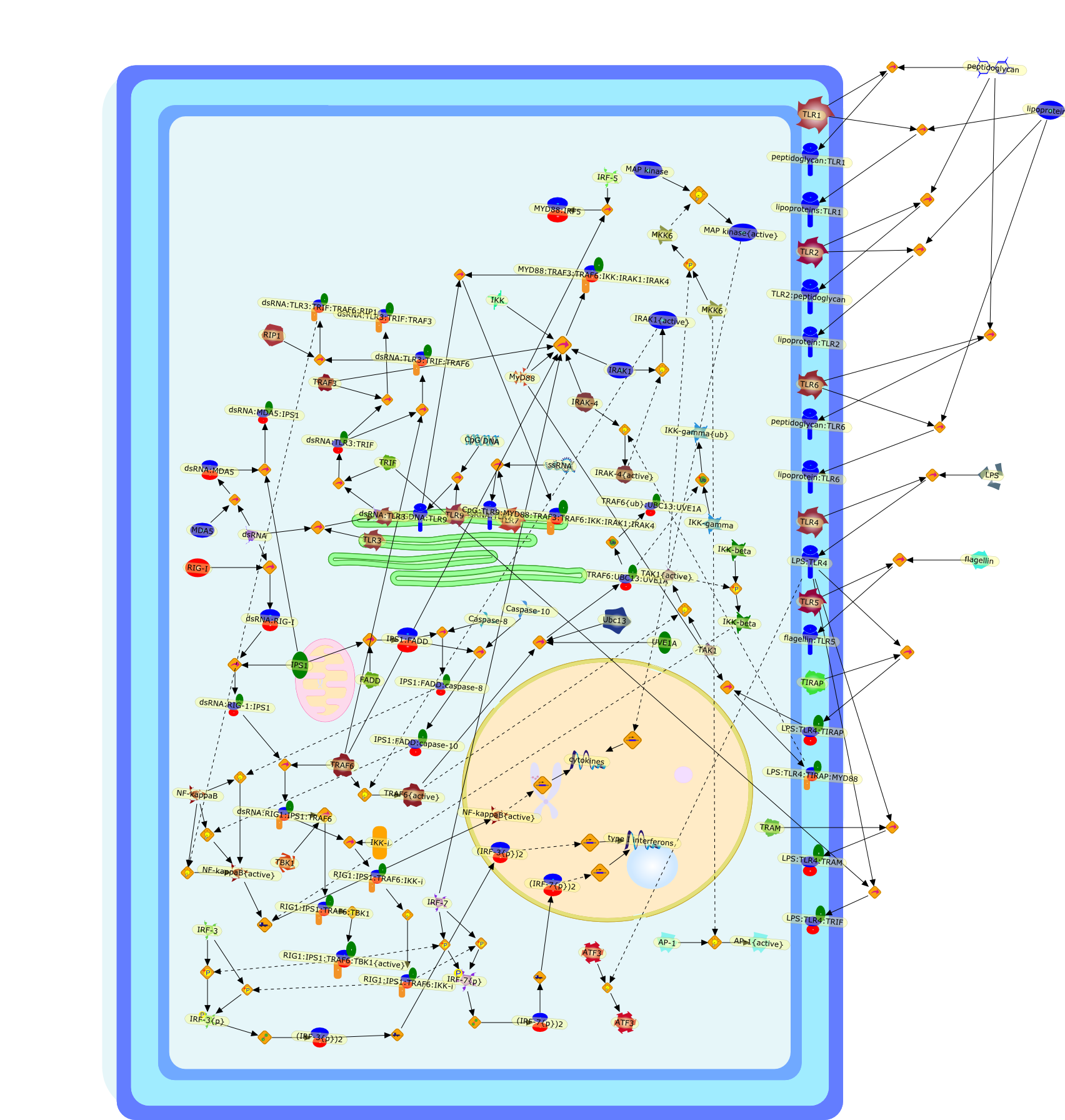
Signaling pathways activated by microorganisms.
Affiliation
Department of Host Defense, Research Institute for Microbial Diseases, OsakaUniversity, ERATO, Japan Science and Technology Agency, 3-1 Yamada-oka, Suita,Osaka 565-0871, Japan.
Abstract
Invasion of viruses and bacteria is initially sensed by the host innate immunesystem, and evokes a rapid inflammatory response. Nucleotides from RNA virusesare recognized by retinoic-acid-inducible gene I-like helicases and Toll-likereceptors, and this recognition triggers signaling cascades that induceantiviral mediators such as type I interferons. By contrast, Toll-like receptorsrecognizing bacterial components induce the expression of proinflammatorycytokines. Furthermore, recent studies suggest that viral and bacterial DNA alsoinduce interferons in a Toll-independent mechanism, possibly throughunidentified cytoplasmic receptor(s).
PMID
17303405
|