| Original Literature | Model OverView |
|---|---|
|
Publication
Title
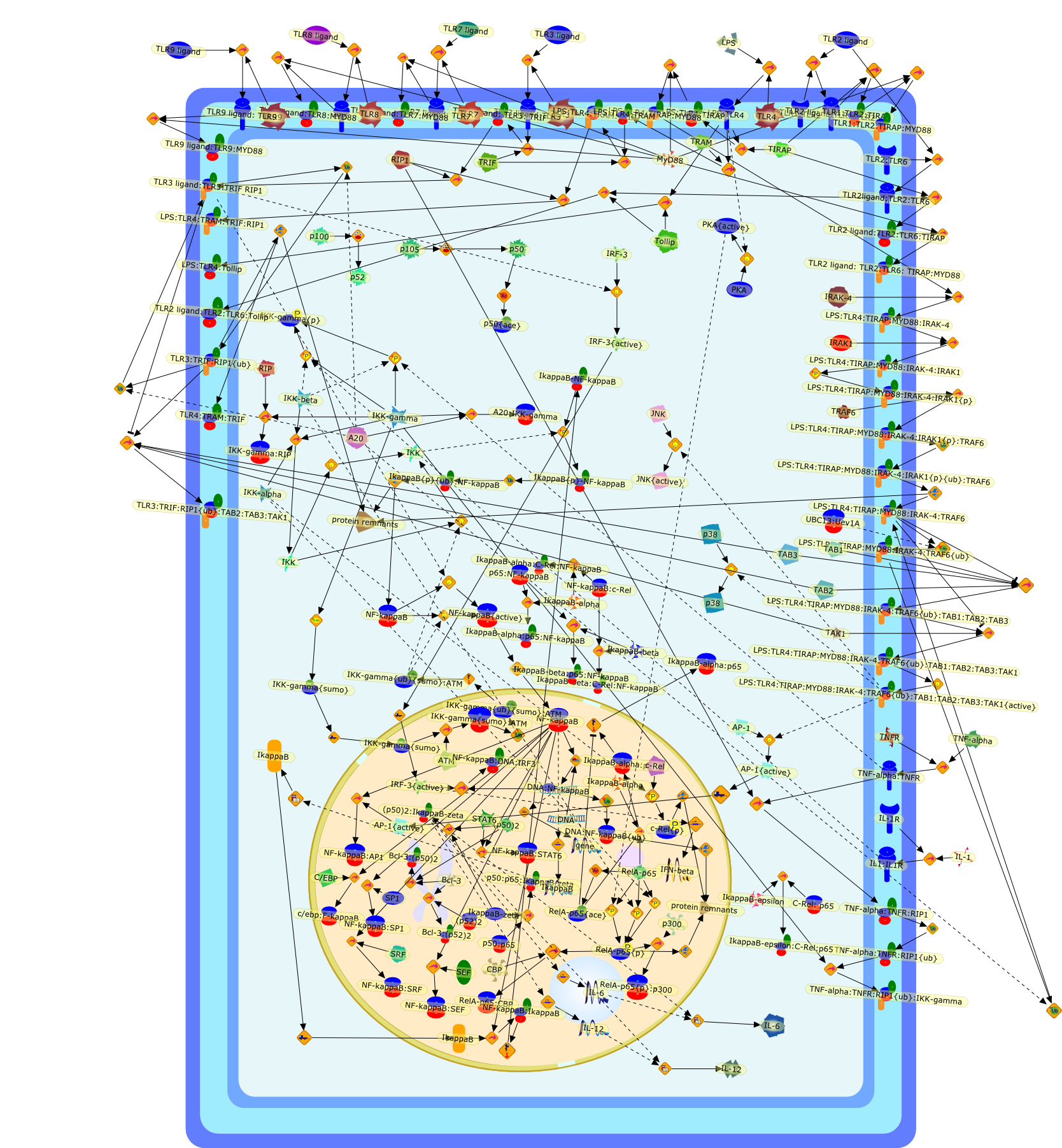
Nuclear factor-kappaB: activation and regulation during toll-like receptorsignaling.
Affiliation
Department of Pathology and Laboratory Medicine, School of Medicine, Universityof Pennsylvania, Philadelphia, PA 19104, USA. rcarmody@mail.med.upenn.edu
Abstract
Toll-like receptors (TLRs) recognize distinct microbial components to initiatethe innate and adaptive immune responses. TLR activation culminates in theexpression of appropriate pro-inflammatory and immunomodulatory factors to meetpathogenic challenges. The transcription factor NF-kappaB is the masterregulator of all TLR-induced responses and its activation is the pivotal eventin TLR-mediated activation of the innate immune response. Many of the keymolecular events required for TLR-induced NF-kappaB activation have beenelucidated. However, much remains to be learned about the ability of TLRs togenerate pathogen-specific responses using a limited number of transcriptionfactors. This review will focus on our current understanding of NF-kappaBactivation by TLRs and potential mechanisms for achieving a signal-specificresponse through NF-kappaB.
PMID
17349209
|