| Original Literature | Model OverView |
|---|---|
|
Publication
Title
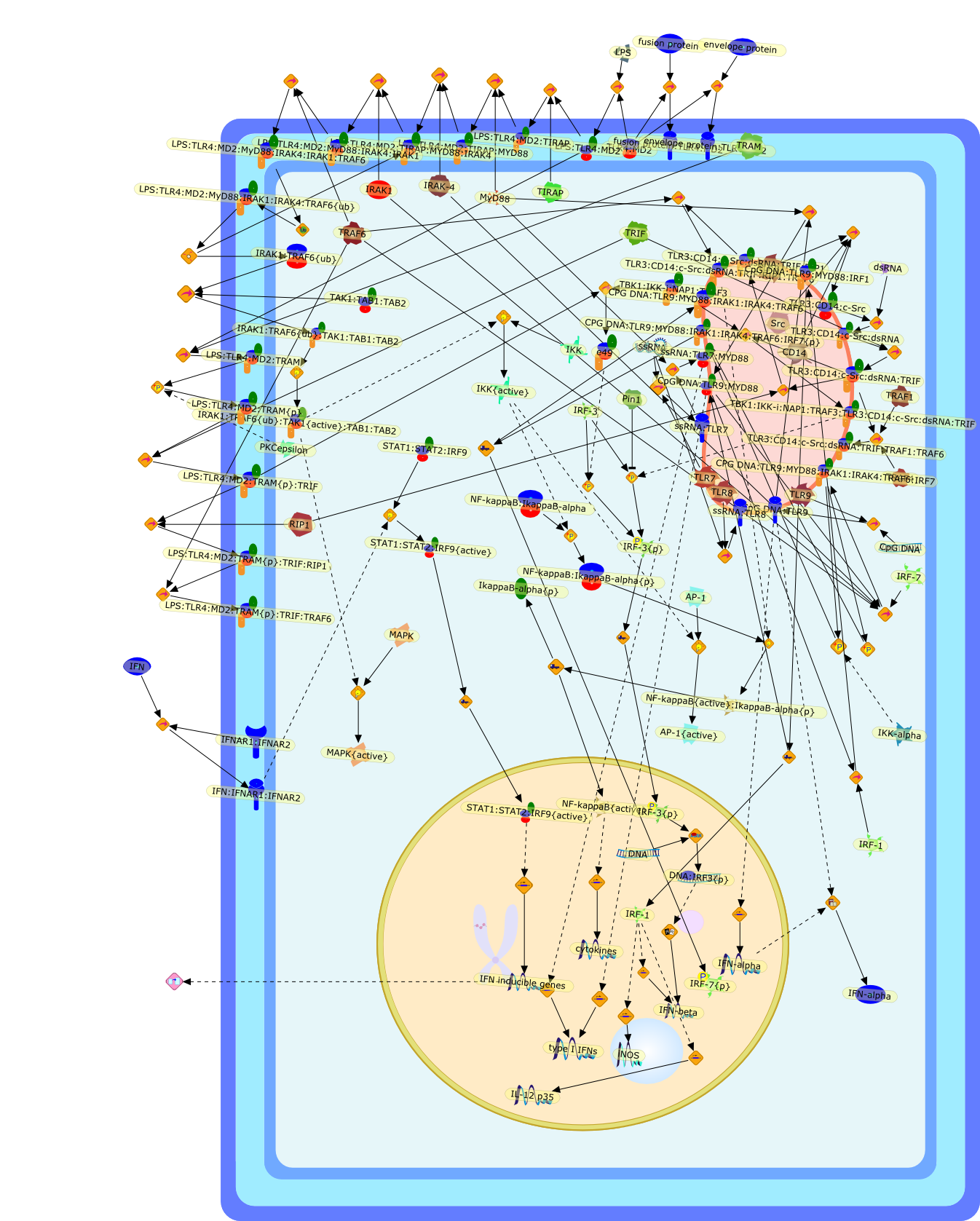
Toll-like receptors and Type I interferons.
Affiliation
Department of Host Defense, Research Institute for Microbial Diseases, OsakaUniversity and ERATO, Japan Science and Technology Corporation, 3-1 Yamada-oka,Suita Osaka 565-0871, Japan.
Abstract
Toll-like receptors (TLRs) are key molecules of the innate immune systems, whichdetect conserved structures found in a broad range of pathogens and triggerinnate immune responses. A subset of TLRs recognizes viral components andinduces antiviral responses. Whereas TLR4 recognizes viral components at thecell surface, TLR3, TLR7, TLR8, and TLR9 recognize viral nucleic acids onendosomal membrane. After ligand recognition, these members activate theirintrinsic signaling pathways and induce type I interferon. In this review, wediscuss the recent findings of the viral recognition by TLRs and their signalingpathways.
PMID
17395581
|