| Original Literature | Model OverView |
|---|---|
|
Publication
Title
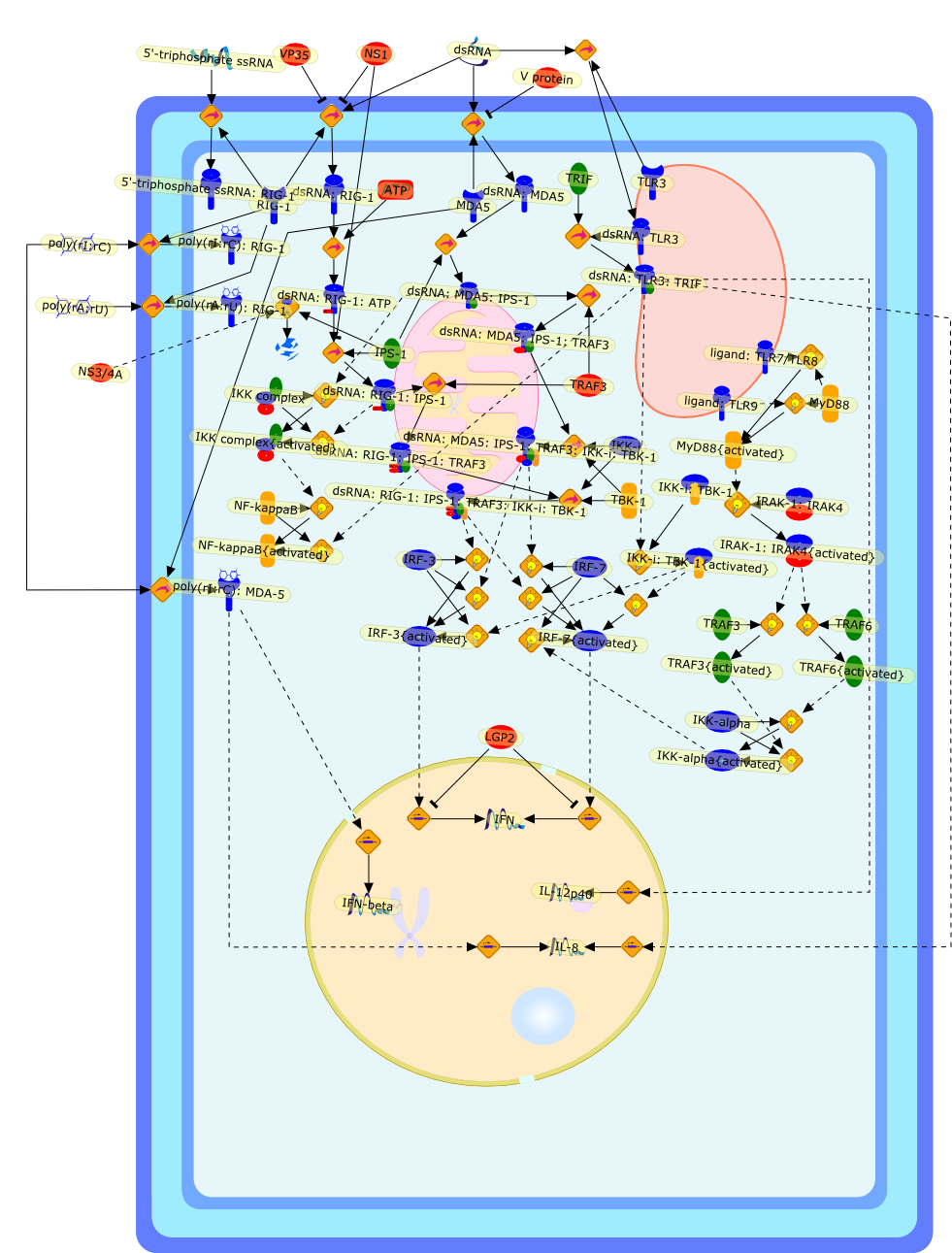
Function of RIG-I-like receptors in antiviral innate immunity.
Affiliation
Laboratory of Molecular Genetics, Institute for Virus Research and GraduateSchool of Biostudies, Kyoto University, Kyoto 606-8507, Japan.
Abstract
PMID
17395582
|