| Original Literature | Model OverView |
|---|---|
|
Publication
Title
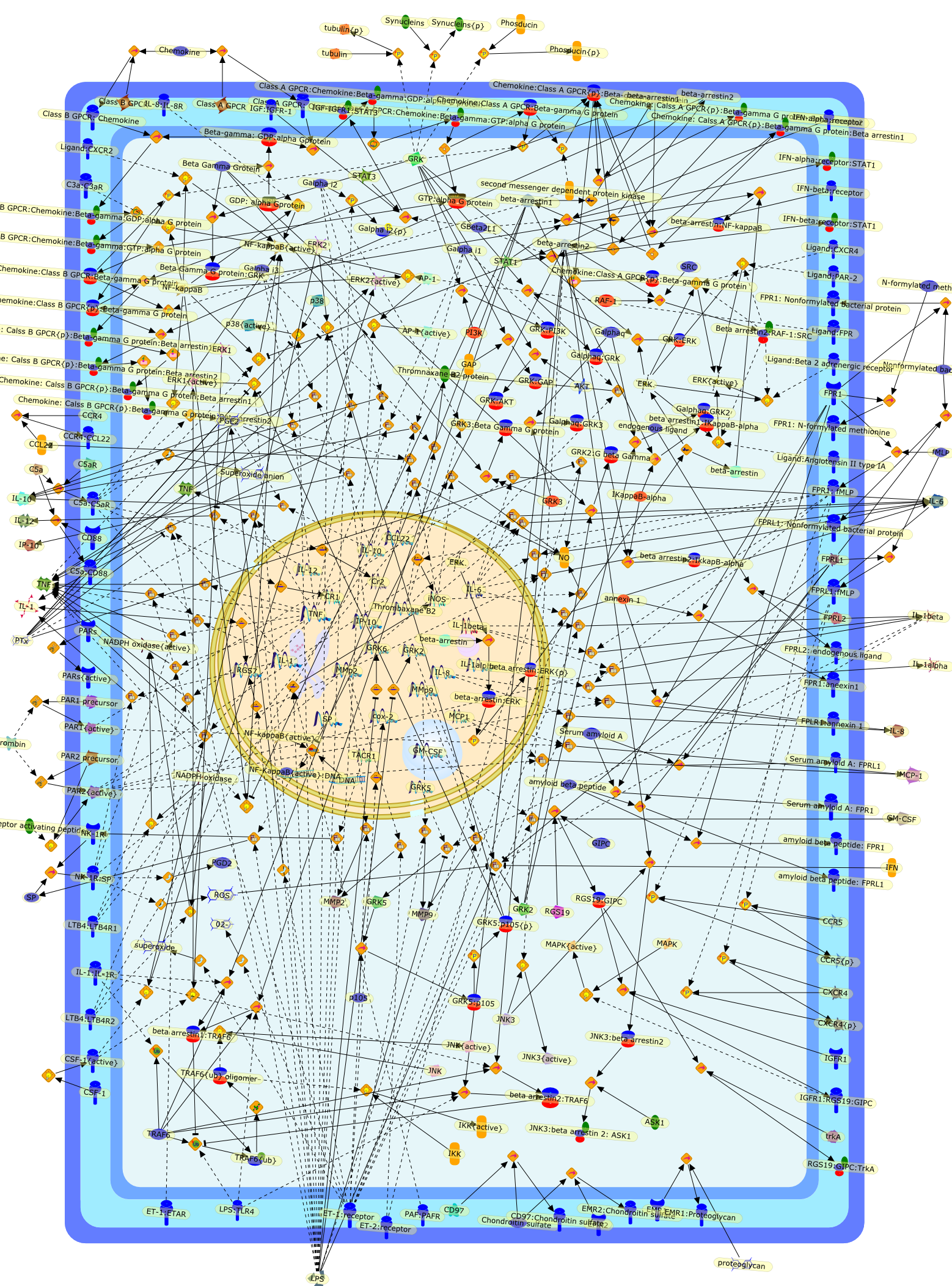
G-protein-coupled receptor expression, function, and signaling in macrophages.
Affiliation
Cooperative Research Centre for Chronic Inflammatory Diseases, University ofQueensland, St. Lucia, Brisbane, QLD 4072, Australia.
Abstract
G-protein-coupled receptors (GPCRs) are widely targeted in drug discovery. Asmacrophages are key cellular mediators of acute and chronic inflammation, wereview here the role of GPCRs in regulating macrophage function, with a focus oncontribution to disease pathology and potential therapeutic applications. Withinthis analysis, we highlight novel GPCRs with a macrophage-restricted expressionprofile, which provide avenues for further exploration. We also review anemerging literature, which documents novel roles for GPCR signaling componentsin GPCR-independent signaling in macrophages. In particular, we examine thecrosstalk between GPCR and TLR signaling pathways and highlight GPCR signalingmolecules which are likely to have uncharacterized functions in this celllineage.
PMID
17456803
|