| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Critical role of toll-like receptors and nucleotide oligomerisation domain inthe regulation of health and disease.
Affiliation
Cardiothoracic Pharmacology, Unit of Critical Care Medicine, Cardiac Medicine,Royal Brompton Hospital, National Heart and Lung Institute, Imperial College,London SW3 6LY, UK. j.a.mitchell@imperial.ac.uk
Abstract
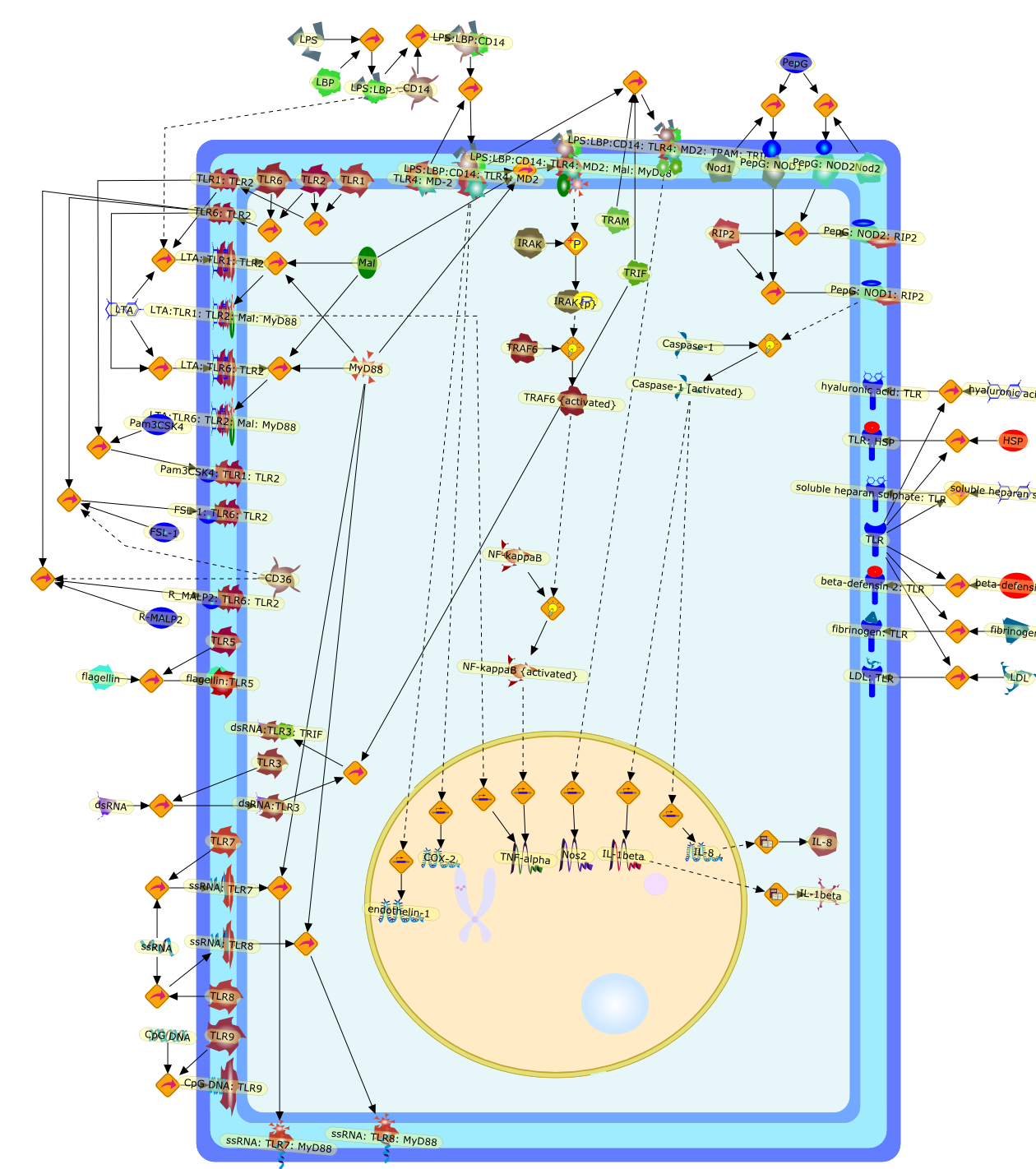
Pathogens are sensed by pattern recognition receptors (PRRs), which are germline-encoded receptors, including transmembrane Toll-like receptors (TLRs) andcytosolic nucleotide oligomerisation domain (NOD) proteins, containingleucine-rich repeats (NLRs). Activation of PRRs by specific pathogen-associatedmolecular patterns (PAMPs) results in genomic responses in host cells involvingactivation transcription factors and the induction of genes. There are now atleast 10 TLRs in humans and 13 in mice, and 2 NLRs (NOD1 and NOD2). TLRsignalling is via interactions with adaptor proteins including MyD88 andtoll-receptor associated activator of interferon (TRIF). NOD signalling is viathe inflammasome and involves activation of Rip-like interactive clarp kinase(RICK). Bacterial lipopolysaccharide (LPS) from Gram-negative bacteria is thebest-studied PAMP and is activated by or 'sensed' by TLR4. Lipoteichoic acid(LTA) from Gram-positive bacteria is sensed by TLR2. TLR4 and TLR2 havedifferent signalling cascades, although activation of either results in symptomsof sepsis and shock. This review describes the rapidly expanding field ofpathogen-sensing receptors and uses LPS and LTA as examples of how thesepathways parallel and diverge from each other. The role of pathogen-sensingpathways in disease is also discussed.
PMID
17535871
|