| Original Literature | Model OverView |
|---|---|
|
Publication
Title
The role of type I interferon production by dendritic cells in host defense.
Affiliation
UMDNJ-New Jersey Medical School, 185 So. Orange Ave., Newark, NJ 07103, USA.bocarsly@umdnj.edu
Abstract
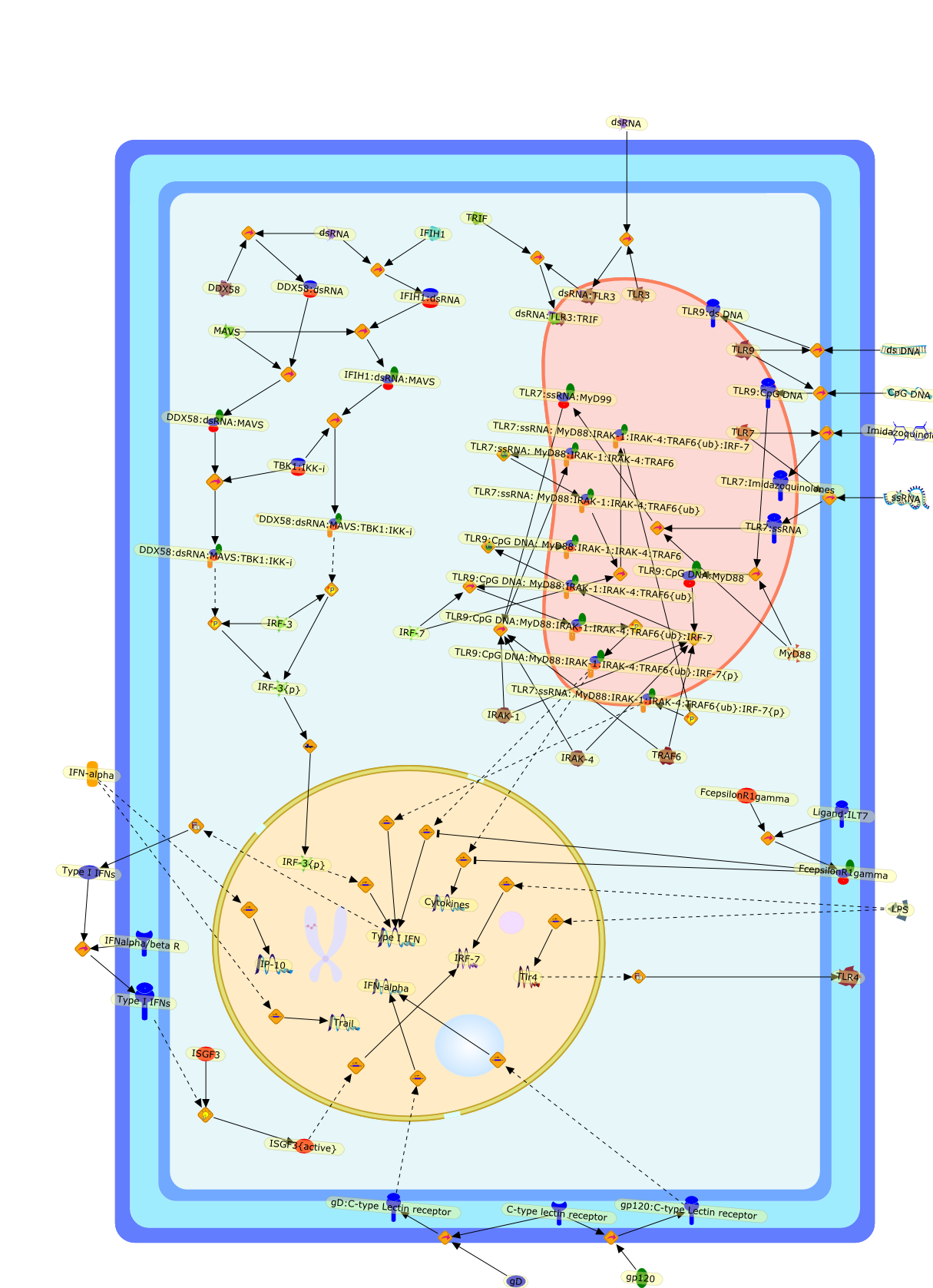
Type I interferons (IFN) and dendritic cells (DC) share an overlapping history,with rapidly accumulating evidence for vital roles for both production of type 1IFN by DC and the interaction of this IFN both with DC and components of theinnate and adaptive immune responses. Within the innate immune response, theplasmacytoid DC (pDC) are the "professional" IFN producing cells, expressingspecialized toll-like receptors (TLR7 and -9) and high constitutive expressionof IRF-7 that allow them to respond to viruses with rapid and extremely robustIFN production; following activation and production of IFN, the pDC subsequentlymature into antigen presenting cells that help to shape the adaptive immuneresponse. However, like most cells in the body, the myeloid or conventional DC(mDC or cDC) also produce type I IFNs, albeit typically at a lower level thanthat observed with pDC, and this IFN is also important in innate and adaptiveimmunity induced by these classic antigen presenting cells. These two major DCsubsets and their IFN products interact both with each other as well as with NKcells, monocytes, T helper cells, T cytotoxic cells, T regulatory cells and Bcells to orchestrate the early immune response. This review discusses some ofthe converging history of DC and IFN as well as mechanisms for IFN induction inDC and the effects of this IFN on the developing immune response.
PMID
17544561
|