| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Pellino proteins: novel players in TLR and IL-1R signalling.
Affiliation
Unit of Molecular Signal Transduction in Inflammation, Department for MolecularBiomedical Research, VIB, Ghent, Belgium.
Abstract
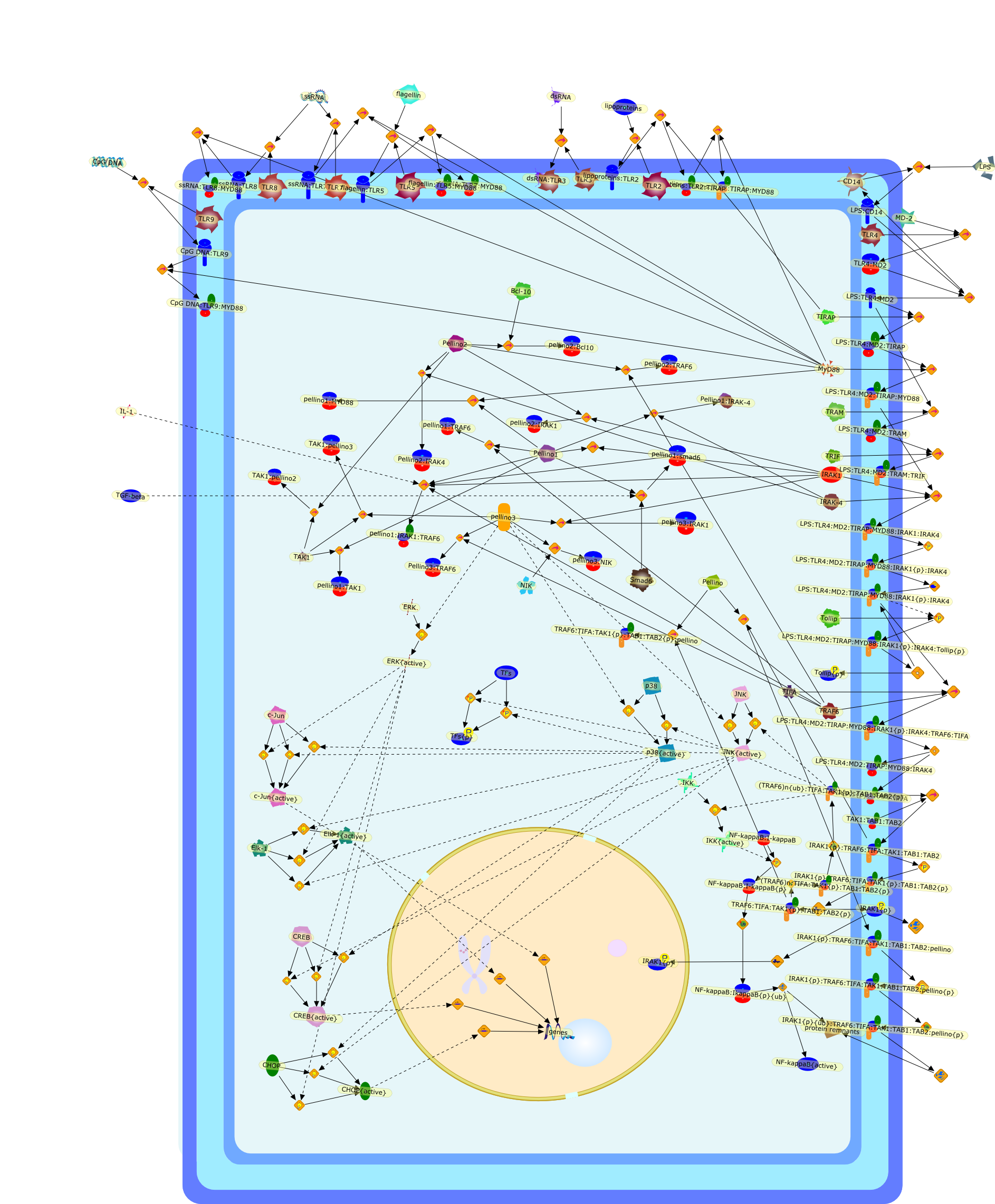
Members of the Toll-like receptor (TLR) and interleukin-1 receptor (IL-1R)family play important roles in immunity and inflammation. They initiate commonintracellular signalling cascades leading to the activation of nuclear factor-?B(NF-?B) and other transcription factors that stimulate the expression of avariety of genes that shape an appropriate immune response. TLR/IL-1R signallinginvolves multiple proteinprotein interactions, but the mechanisms that regulatethese interactions are still largely unclear. In this context, Pellino proteinshave been suggested to function as evolutionary conserved scaffold proteins inTLR/IL-1R signalling. However, recently Pellino proteins were also proposed tofunction as novel ubiquitin ligases for IL-1R associated kinase 1 (IRAK-1). Herewe review our current knowledge on the expression, biological role and mechanismof action of Pellino proteins in TLR/IL-1R-induced signalling.
PMID
17635639
|