| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins.
Affiliation
School of Biochemistry and Immunology, Trinity College, Dublin, Ireland.wattert@tcd.ie
Abstract
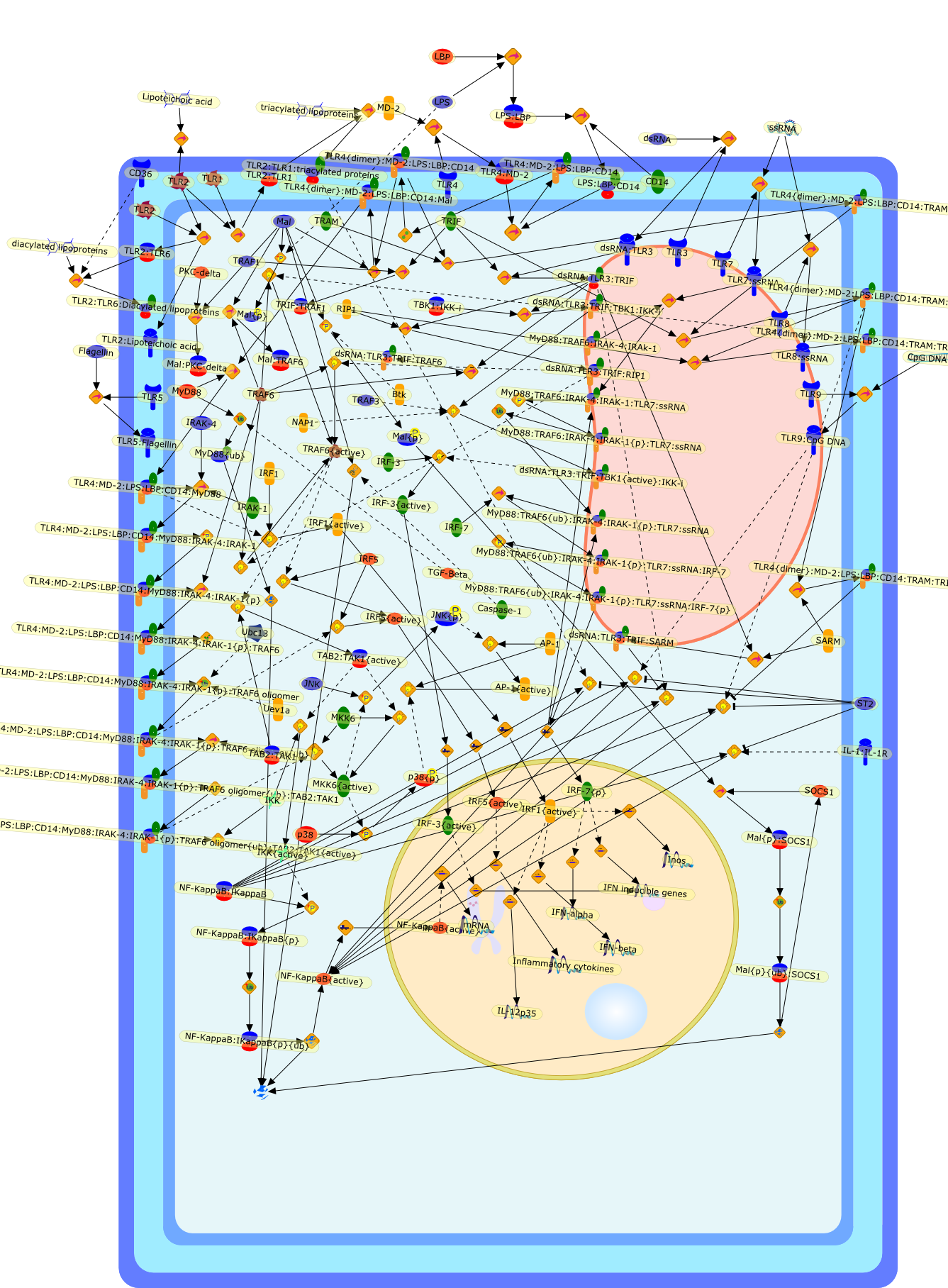
The Toll/IL-1 receptor (TIR) domain plays a central role in Toll-like receptor(TLR) signalling. All TLRs contain a cytoplasmic TIR domain, which, uponactivation, acts as a scaffold to recruit adaptor proteins. The adaptor proteinsMyD88, Mal, TRIF, TRAM and SARM are also characterized by the presence of a TIRdomain. MyD88, Mal, TRIF and TRAM associate with the TLRs via homophilic TIRdomain interactions whereas SARM utilizes its TIR domain to negatively regulateTRIF. It is well established that the differential recruitment of adaptors toTLRs provides a significant amount of specificity to the TLR-signallingpathways. Despite this, the TIR-TIR interface has not been well defined.However, structural studies have indicated the importance of TIR domain surfacesin mediating specific TIR-TIR interactions. Furthermore, recent findingsregarding the regulation of adaptors provide further insight into the crucialrole of the TIR domain in TLR signalling.
PMID
17667936
|