| Original Literature | Model OverView |
|---|---|
|
Publication
Title
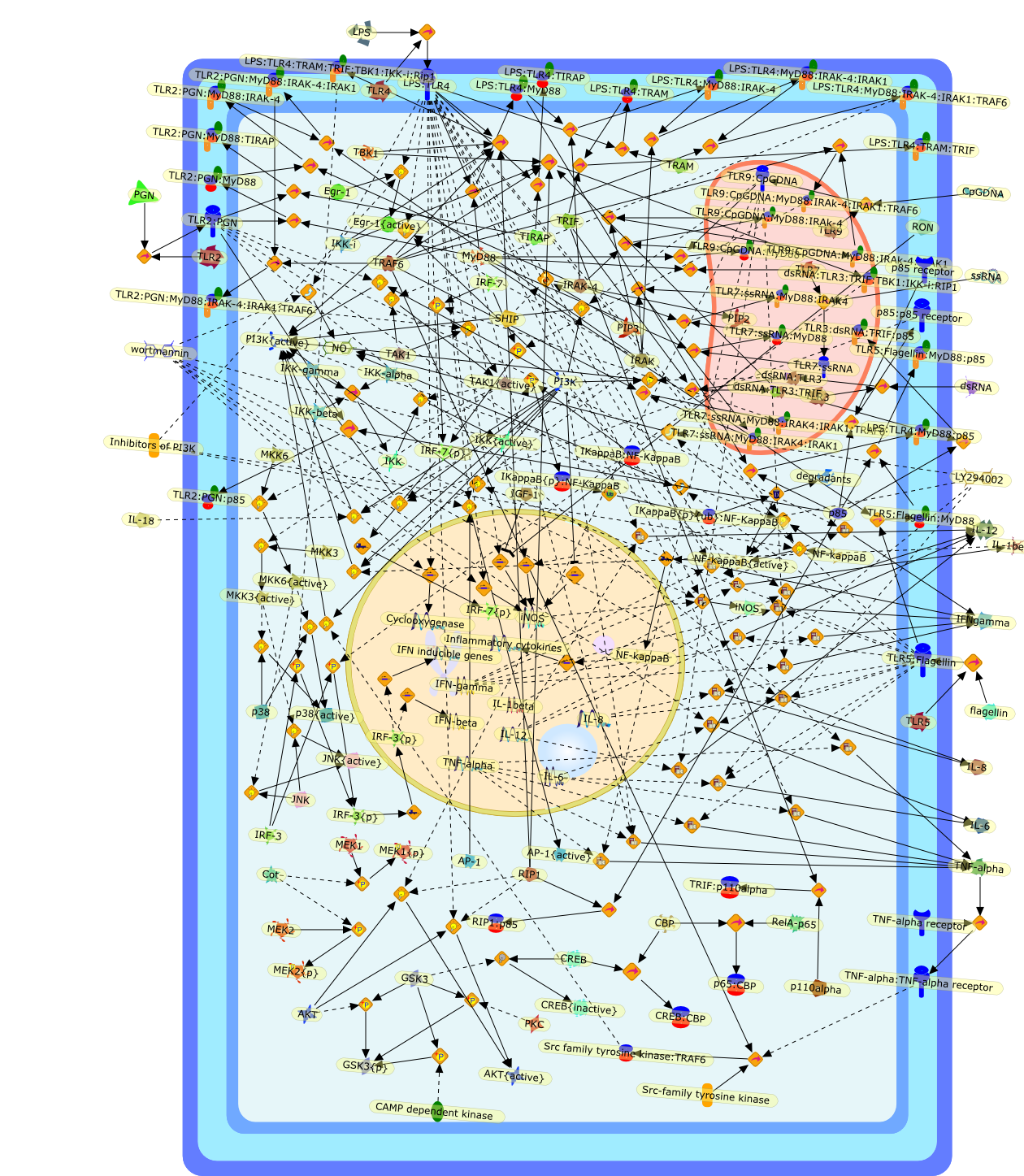
Role of phosphoinositide 3-kinase in innate immunity.
Affiliation
Division of Molecular Medical Science, Graduate School of Biomedical Sciences,Hiroshima University, Hiroshima, Japan. khazeki@hiroshima-u.ac.jp
Abstract
Recent advances in our understanding of the molecular basis of mammalian hostimmune responses to microbial invasion suggest that the first line of defenseagainst microbes is the recognition of pathogen-associated molecular patterns byToll-like receptors (TLRs). Phosphoinositide 3-kinase (PI3K) is thought toparticipate in the TLR signaling pathway. The activation of PI3K is commonlyobserved after stimulation with various TLR ligands. The resultant activation ofa serine-threonine protein kinase Akt leads to the phosphorylation of glycogensynthase kinase (GSK)-3beta, which is active in resting cells but is inactivatedby phosphorylation. GSK-3beta has been linked to the regulation of a multitudeof transcription factors, including NF-kappaB, AP-1, NF-AT, and CREB eithernegatively or positively. Thus, the altered activity of GSK-3beta causes diverseeffects on cytokine expression. Generally, activation of PI3K results in theinhibition of proinflammatory events such as expression of IL-12 and TNF-alpha.Thus, PI3K is a negative regulator of TLR signaling. Among the members of theClass I PI3K family, p85/p110beta appears to be the subtype activated on TLRligation, but the molecular basis for this specificity has yet to be elucidated.
PMID
17827709
|