| Original Literature | Model OverView |
|---|---|
|
Publication
Title
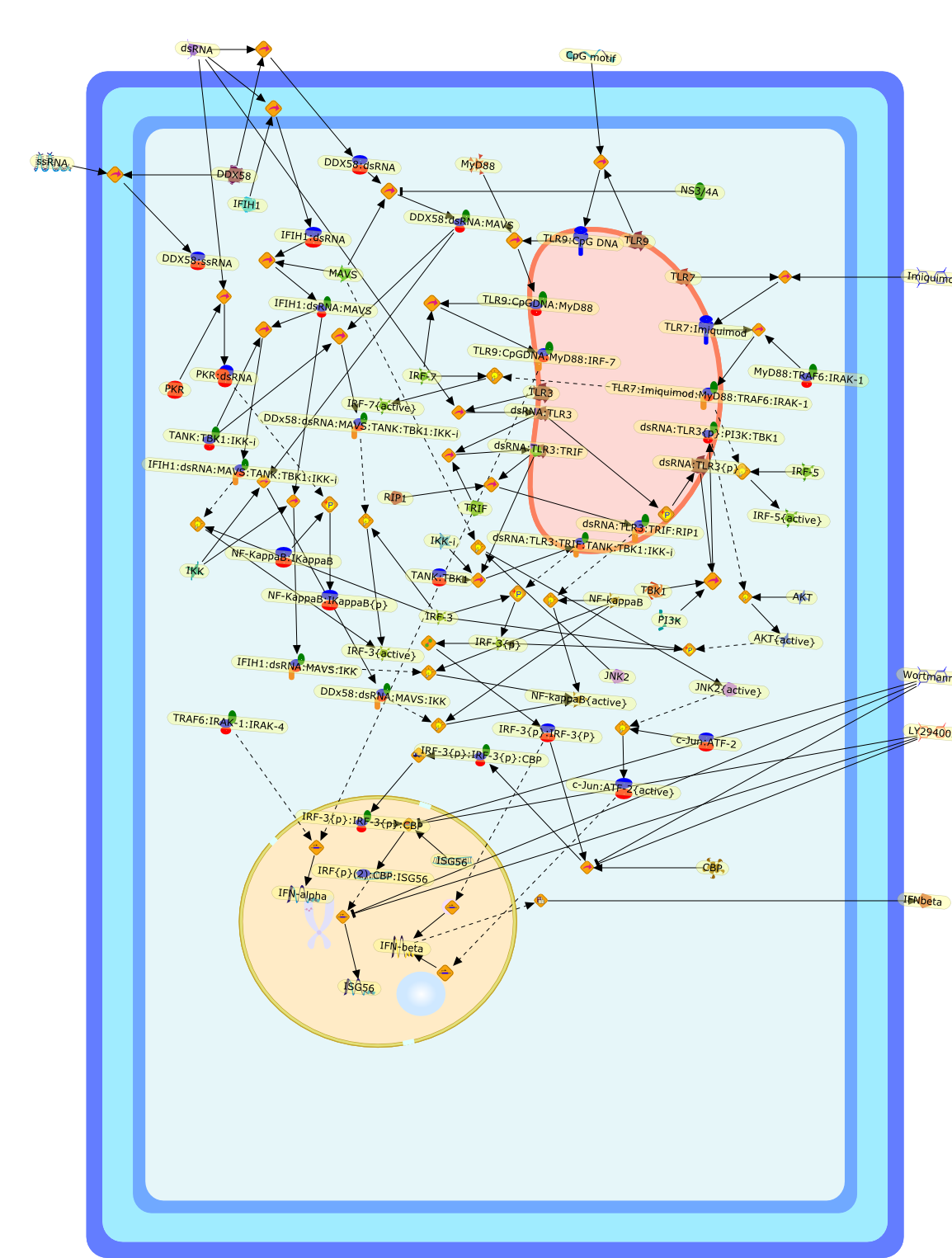
Signalling pathways mediating type I interferon gene expression.
Affiliation
Department of Respiratory Medicine, National Heart and Lung Institute, MRCCentre in Allergic Mechanisms of Asthma, St Marys Campus, Norfolk Place, LondonW2 1PG, UK. michael.edwards@imperial.ac.uk
Abstract
Type I interferon-alpha/beta play an essential role in immunity to viruses.While interferon-beta has been used as a model of a complex promoter, many ofthe signalling pathways leading to interferon-beta gene expression remaincontroversial. Recent milestones include the discovery of Toll-like receptorsand RNA helicases that signal via a novel kinase complex composed of I kappa Bkinase-iota/epsilon or TANK binding kinase-1. This review provides a timelysummary of this rapidly expanding field, focusing specifically on the variousviral RNA binding molecules and their associated signalling pathways.
PMID
17904888
|