| Original Literature | Model OverView |
|---|---|
|
Publication
Title
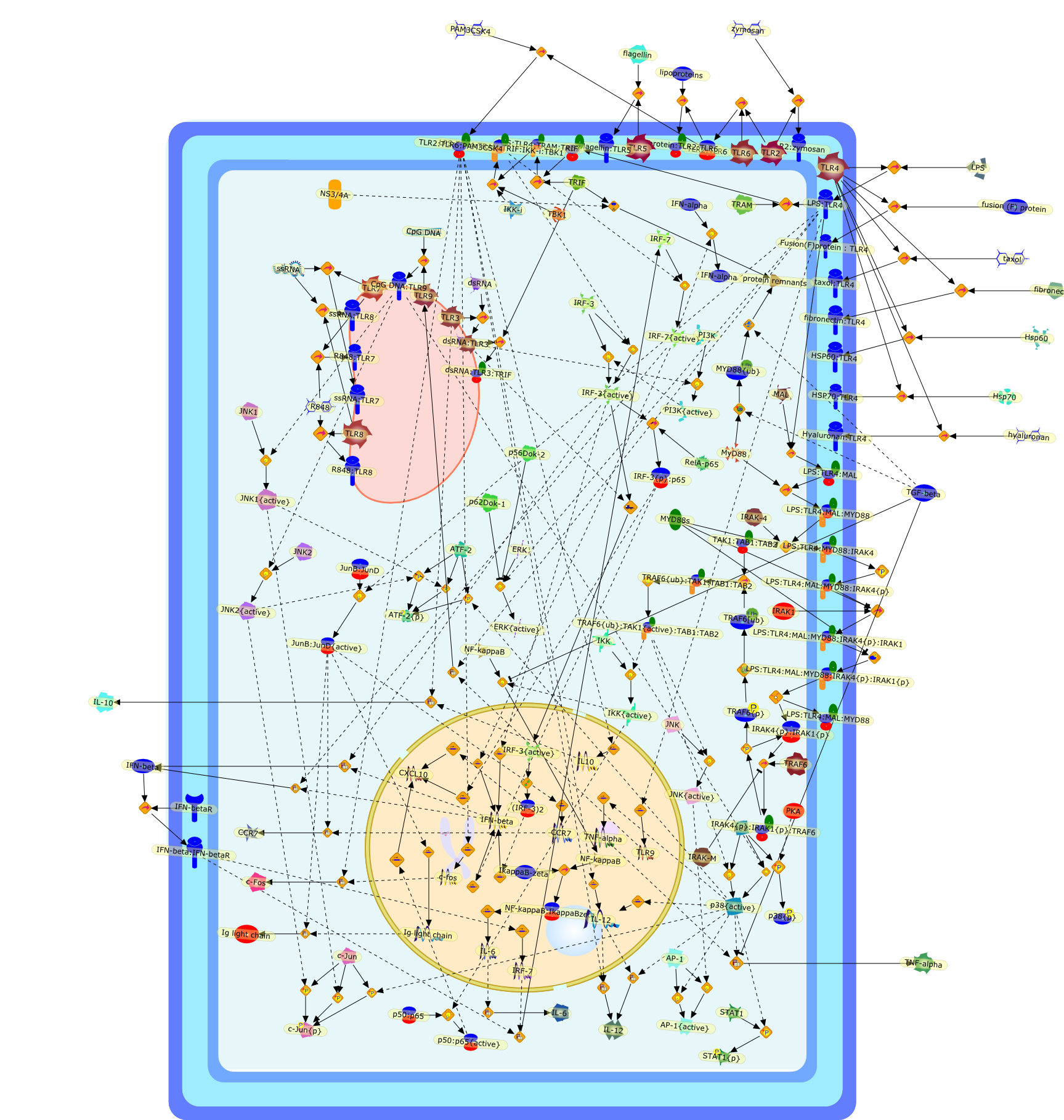
Toll-like receptor signal transduction.
Affiliation
Department of Molecular Science and Technology, Ajou University, Suwon 443-749,Korea.
Abstract
Toll-like receptors (TLRs) are the archetypal pattern recognition receptors insensing exogenous pathogens. Activation of TLRs is a first line of defense ofthe immune system, leading to the activation and recruitment of neutrophils andmacrophages to sites of infection and enhances antimicrobial activity. The TLRsignaling through different intracellular molecules, such as MAP kinases andIkappaB kinases which are conserved signaling elements for many receptors, leadsto a distinct set of proinflammatory gene expressions. However, how thesepathways differentially and precisely control the transcription of identicalgenes remains largely unknown. Our review focuses on the details of up-to-datesignaling molecules including negative regulators and their role in controllinginnate immune response. We also stress the importance of developing systemicapproaches for the global understanding of TLR signaling so that appropriatedrug therapeutic targets can be identified for regulating inflammatory diseases.
PMID
17934330
|