| Original Literature | Model OverView |
|---|---|
|
Publication
Title
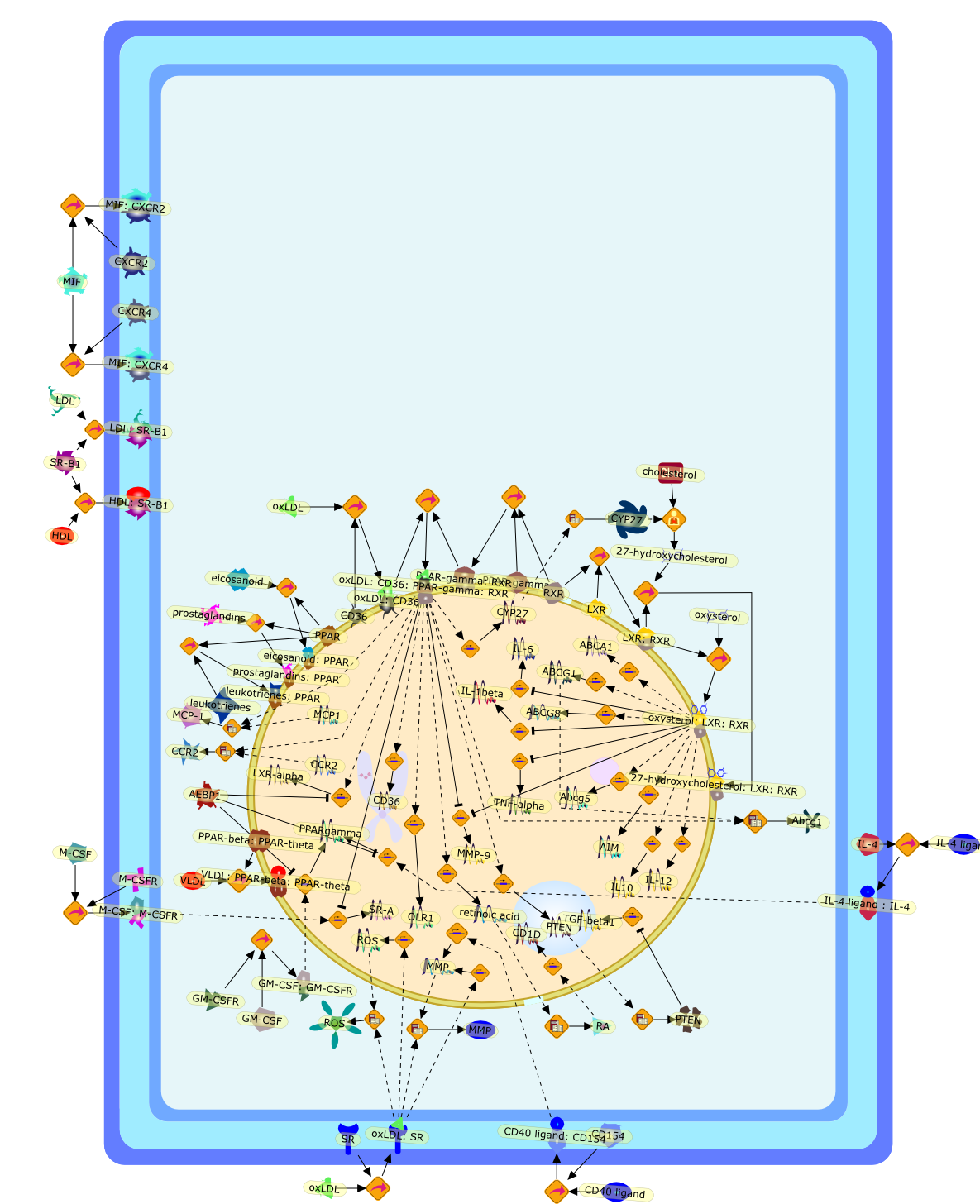
Nuclear receptors in macrophages: a link between metabolism and inflammation.
Affiliation
University of Debrecen, Medical and Health Science Center, Research Center forMolecular Medicine, Life Science Building, Department of Biochemistry andMolecular Biology, Debrecen, Egyetem ter 1 H-4032, Hungary. szantoa@dote.hu
Abstract
Subclinical inflammation is a candidate etiological factor in the pathogenesisof metabolic syndrome and in the progression of atherosclerosis. A central rolefor activated macrophages has been elucidated recently as important regulatorsof the inflammatory process in atherosclerosis. Macrophage differentiation andfunction can be modulated by a class of transcription factors termed nuclearreceptors. These are activated by intermediary products of basic metabolicprocesses. In this review the contribution of peroxisome proliferator-activatedreceptors and liver X receptors to macrophage functions in inflammation andlipid metabolism will be discussed in light of their roles in macrophages duringatherosclerosis. In the past decade much effort has been made to understand themechanisms how lipids are handled by macrophages and how inflammation couldpromote the atherogenic process. Here, we also provide an overview of these twofields.
PMID
18022390
|