| Original Literature | Model OverView |
|---|---|
|
Publication
Title
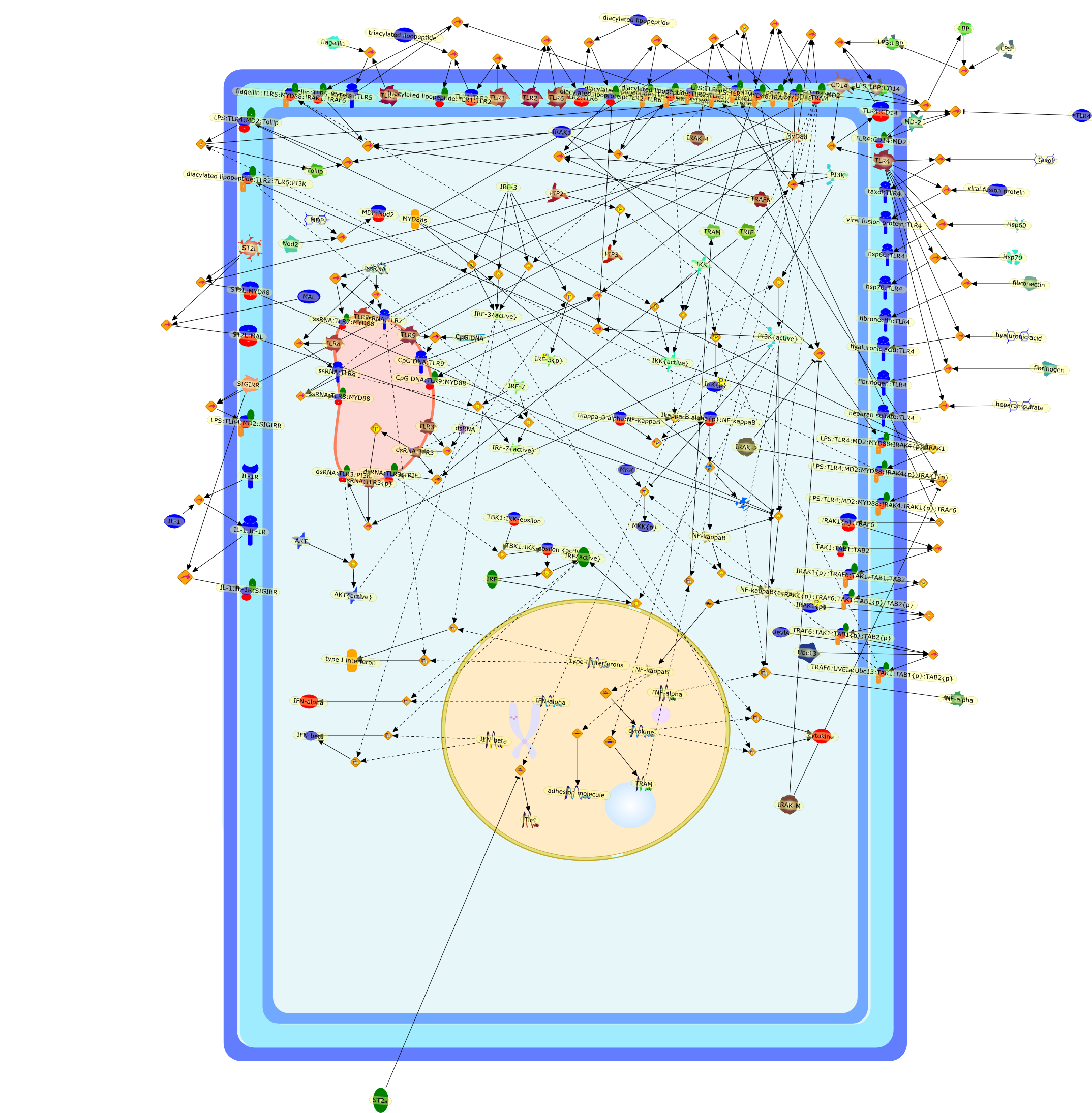
Toll-like receptors are key participants in innate immune responses.
Affiliation
Programa Disciplinario de Inmunologia, Instituto de Ciencias Biomedicas,Facultad de Medicina, Universidad de Chile, Santiago, Chile.
Abstract
During an infection, one of the principal challenges for the host is to detectthe pathogen and activate a rapid defensive response. The Toll-like family ofreceptors (TLRs), among other pattern recognition receptors (PRR), performs thisdetection process in vertebrate and invertebrate organisms. These type Itransmembrane receptors identify microbial conserved structures orpathogen-associated molecular patterns (PAMPs). Recognition of microbialcomponents by TLRs initiates signaling transduction pathways that induce geneexpression. These gene products regulate innate immune responses and furtherdevelop an antigen-specific acquired immunity. TLR signaling pathways areregulated by intracellular adaptor molecules, such as MyD88, TIRAP/Mal, betweenothers that provide specificity of individual TLR- mediated signaling pathways.TLR-mediated activation of innate immunity is involved not only in host defenseagainst pathogens but also in immune disorders. The involvement of TLR-mediatedpathways in auto-immune and inflammatory diseases is described in this reviewarticle.
PMID
18064347
|