| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Role of Toll-like receptor responses for sepsis pathogenesis.
Affiliation
Department of Surgery, Technische Universitat Munchen, Ismaninger Strasse 22,81675, Munich, Germany. Heike.Weighardt@uni-duesseldorf.de
Abstract
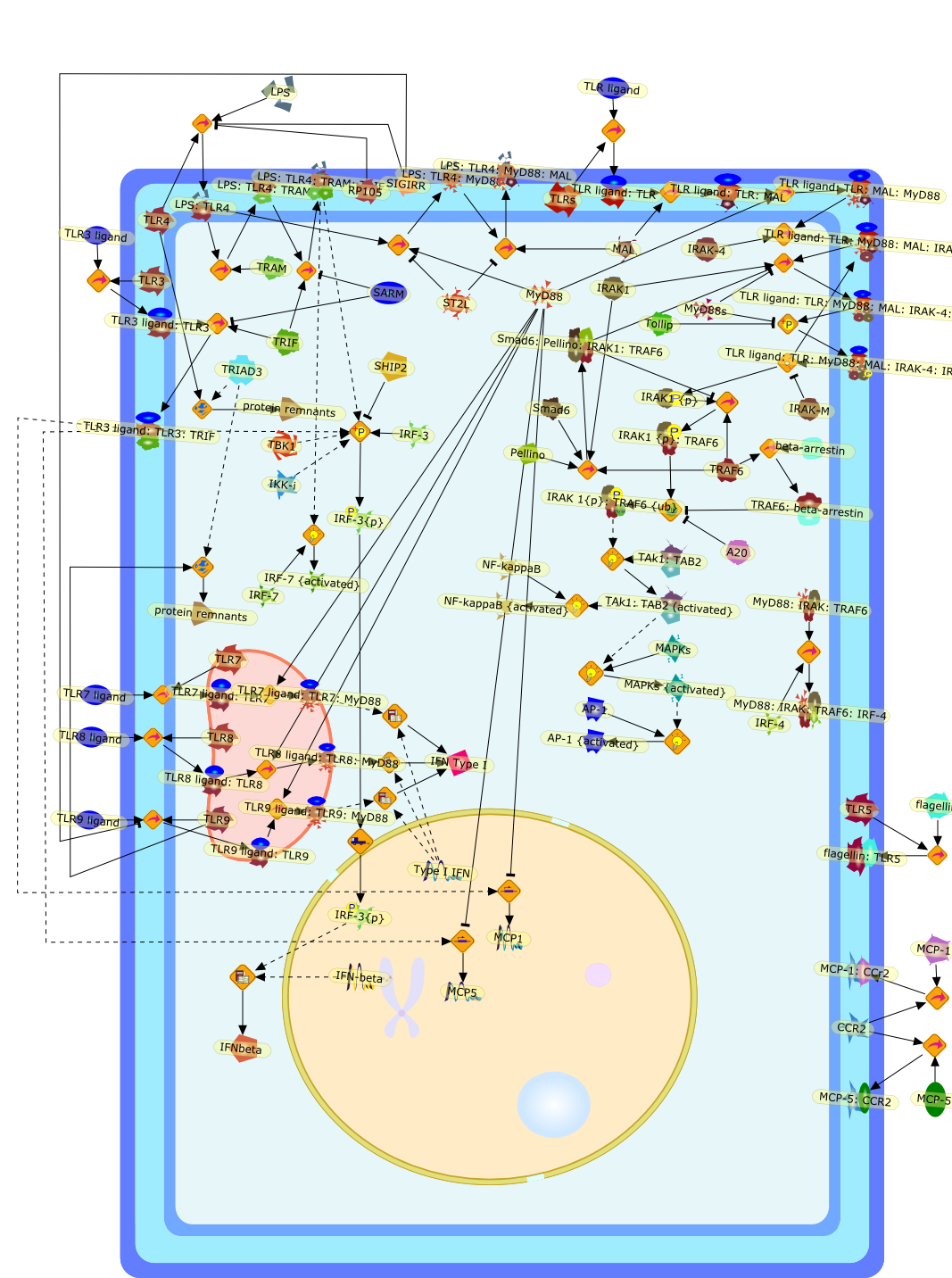
Sepsis remains a serious clinical problem because of high patient morbidity andmortality. Despite significant advances in critical care, there is still noefficient causal therapy applicable to patients indicating the need to furtherelucidate the molecular pathways leading to the immunopathology of sepsis. Theimportance of Toll-like receptors (TLR) for the induction of immune responsesagainst sepsis was demonstrated in humans exhibiting polymorphisms in TLR genesand in animal models using genetically modified mouse strains. Because of theclinical heterogeneity in human sepsis and the complex pathomechanismsunderlying sepsis, several different animal models might be used to cover thediverse features of sepsis. TLR receptors induce signaling through the adapterproteins MyD88 and TRIF. TLR signaling is tightly controlled at different stepsof the signaling cascade by series of regulatory proteins. Using a model ofsevere polymicrobial septic peritonitis we could show that single TLRs aredispensable for the induction of innate immune responses under those conditions.However, genetic ablation of MyD88 or TRIF/type-I interferon signaling pathwaysprevented hyper-inflammation and attenuated the pathogenic consequences ofsepsis indicating that dampening common signaling pathways may create a moderatesignal strength which is associated with favorable immune responses. Therefore,broad knowledge about the regulation of TLR-induced signaling pathways mayfurther elucidate the immune mechanisms during sepsis and targeting of TLRadapter molecules may provide a new therapeutic strategy against severe sepsis.
PMID
18086373
|