| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Molecular mechanisms of the anti-inflammatory functions of interferons.
Affiliation
Max F Perutz Laboratories, University of Vienna, Dr Bohr-Gasse 9, A-1030,Vienna, Austria. pavel.kovarik@univie.ac.at
Abstract
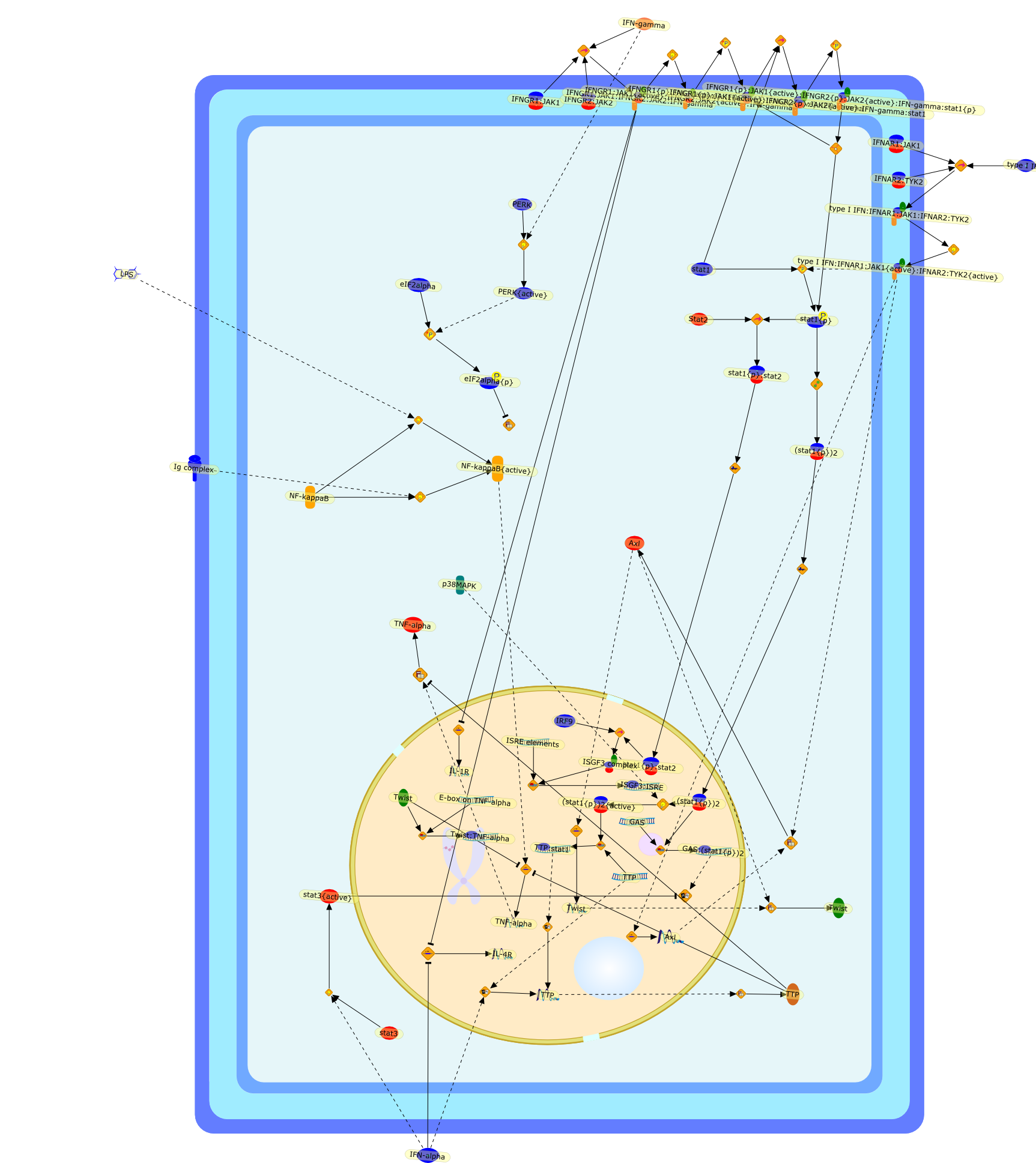
Interferons are pleiotropic cytokines with important proinflammatory functionsrequired in defence against infections with bacteria, viruses and multicellularparasites. In recent years, fundamental functions of interferons in otherprocesses such as cancer immunosurveillance, immune homeostasis andimmunosuppression have been established. In addition, anti-inflammatory roles ofinterferons are well-documented in several inflammatory disease models in themouse, most importantly in experimental autoimmune encephalomyelitis thatresembles multiple sclerosis in humans. While the beneficial effects ofinterferons in such disease models are known, the molecular mechanisms remainpoorly understood. Only recently a few molecular principles for theanti-inflammatory properties of interferons at the cellular level have beenrevealed. They include the ability of interferons to reduce the expression ofthe receptors for the inflammation-related cytokines IL-1 and IL-4, or toincrease the expression of the potent anti-inflammatory genes tristetraprolinand Twist. However, the individual contribution of these anti-inflammatoryresponses to the overall beneficial effects of interferons in inflammatorydiseases is still an open question. Also, the reason for the apparently limitednumber of tissues that are susceptible to the anti-inflammatory functions ofinterferons remains enigmatic. This review summarizes the present knowledge ofthe anti-inflammatory effects of interferons, and describes the currently knownmolecular mechanisms that may help explain the benefits of interferon signallingin several inflammatory diseases.
PMID
18086388
|