| Original Literature | Model OverView |
|---|---|
|
Publication
Title
C-type lectin receptors in antifungal immunity.
Affiliation
Institute of Infectious Disease and Molecular Medicine, Division of Immunology,CLS, University of Cape Town, Observatory, Cape Town, 7925, South Africa.
Abstract
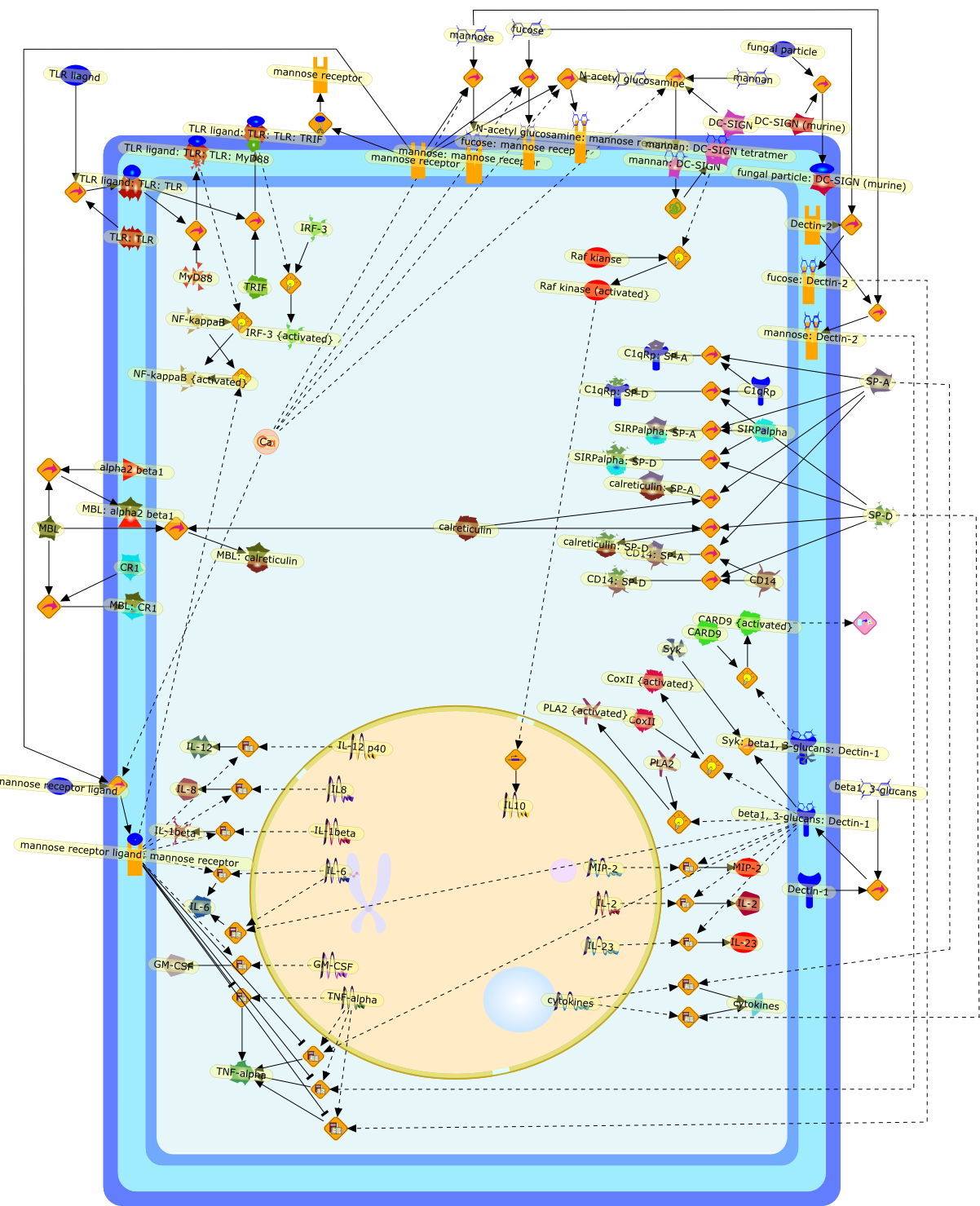
Fungal infections represent a significant health burden, especially inimmunocompromised individuals, yet many of the underlying immunologicalmechanisms involved in the recognition and control of these pathogens areunclear. The identification of the Toll-like receptors (TLRs) has shed newinsights on innate microbial recognition and the initiation of immune responses;however, recent evidence indicates that the 'non-TLR' receptors also have asignificant role in these processes, particularly in antifungal immunity. Ofinterest are members of the C-type lectin-receptor family, including the mannosereceptor, dendritic cell-specific intercellular adhesion molecule-3(ICAM-3)-grabbing non-integrin (DC-SIGN), Dectin-1, Dectin-2 and the collectins.Here, we review the roles of each of these receptors, describing how theycontribute to fungal recognition, uptake and killing and also participate in theinduction and/or modulation of the host immune response.
PMID
18160296
|