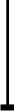| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Macrophage activation by endogenous danger signals.
Affiliation
Department of Cell Biology and Molecular Genetics, University of Maryland,College Park, MD 20742, USA.
Abstract
Macrophages are cells that function as a first line of defence against invadingmicroorganisms. One of the hallmarks of macrophages is their ability to becomeactivated in response to exogenous 'danger signals'. Most microbes havemolecular patterns (PAMPS) that are recognized by macrophages and trigger thisactivation response. There are many aspects of the activation response to PAMPSthat are recapitulated when macrophages encounter endogenous danger signals. Inresponse to damaged or stressed self, macrophages undergo physiological changesthat include the initiation of signal transduction cascades fromgermline-encoded receptors, resulting in the elaboration of chemokines,cytokines and toxic mediators. This response to endogenous mediators can enhanceinflammation, and thereby contribute to autoimmune pathologies. Often theoverall inflammatory response is the result of cooperative activation signalsfrom both exogenous and endogenous signals. Macrophage activation plays acritical role, not only in the initiation of the inflammatory response but alsoin the resolution of this response. The clearance of granulocytes and theelaboration of anti-inflammatory mediators by macrophages contribute to thedissolution of the inflammatory response. Thus, macrophages are a key player inthe initiation, propagation and resolution of inflammation. This reviewsummarizes our understanding of the role of macrophages in inflammation. We payparticular attention to the endogenous danger signals that macrophages mayencounter and the responses that these signals induce. The molecular mechanismsresponsible for these responses and the diseases that result frominappropriately controlled macrophage activation are also examined. 2007Pathological Society of Great Britain and Ireland
PMID
18161744
|