| Original Literature | Model OverView |
|---|---|
|
Publication
Title
MyDths and un-TOLLed truths: sensor, instructive and effector immunity totuberculosis.
Affiliation
Microbial Interface Biology, Research Center Borstel, D-23845 Borstel, Germany.
Abstract
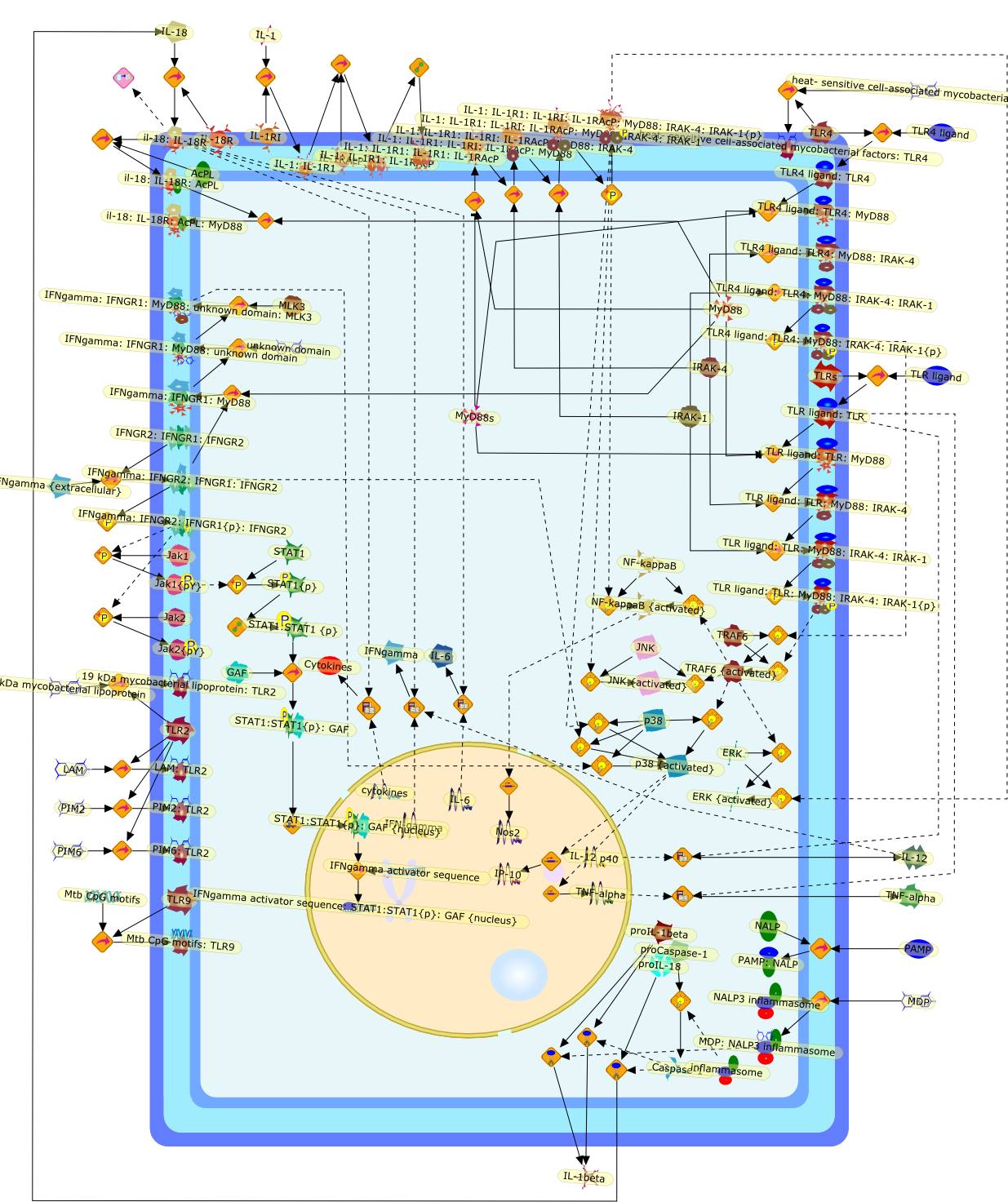
Controversy exists concerning the role of Toll-like receptors and MyD88 inimmunity to tuberculosis (TB). This mini-review argues that (i) Toll-likereceptors are not essential for an effective immune response against TB, (ii)MyD88 is essential, but not because it transduces signals from TLRs, (iii)adaptive immunity to TB is largely TLR/MyD88-independent. Some of thediscrepancies may be resolved by cogent attribution of distinct immune functionsto the individual components of the TLR/MyD88 system. In mice, TLRs and MyD88are fully dispensable in sensing Mtb infection and instructing T cell-mediatedadaptive immunity, and while TLRs are also redundant during macrophage effectorimmunity, MyD88 is essential for efficient killing of mycobacteria. Thisdistinction should help to molecularly pinpoint the MyD88-dependent, yetTLR-independent critical mechanisms of macrophage activation involved inintracellular growth restriction of Mtb. Disrupted IL-1R and/or IFN-gammasignaling pathways likely play a much more prominent role in explaining theexquisite susceptibility of MyD88-deficient mice to TB than the function ofMyD88 as a TLR adaptor.
PMID
18191460
|