| Original Literature | Model OverView |
|---|---|
|
Publication
Title
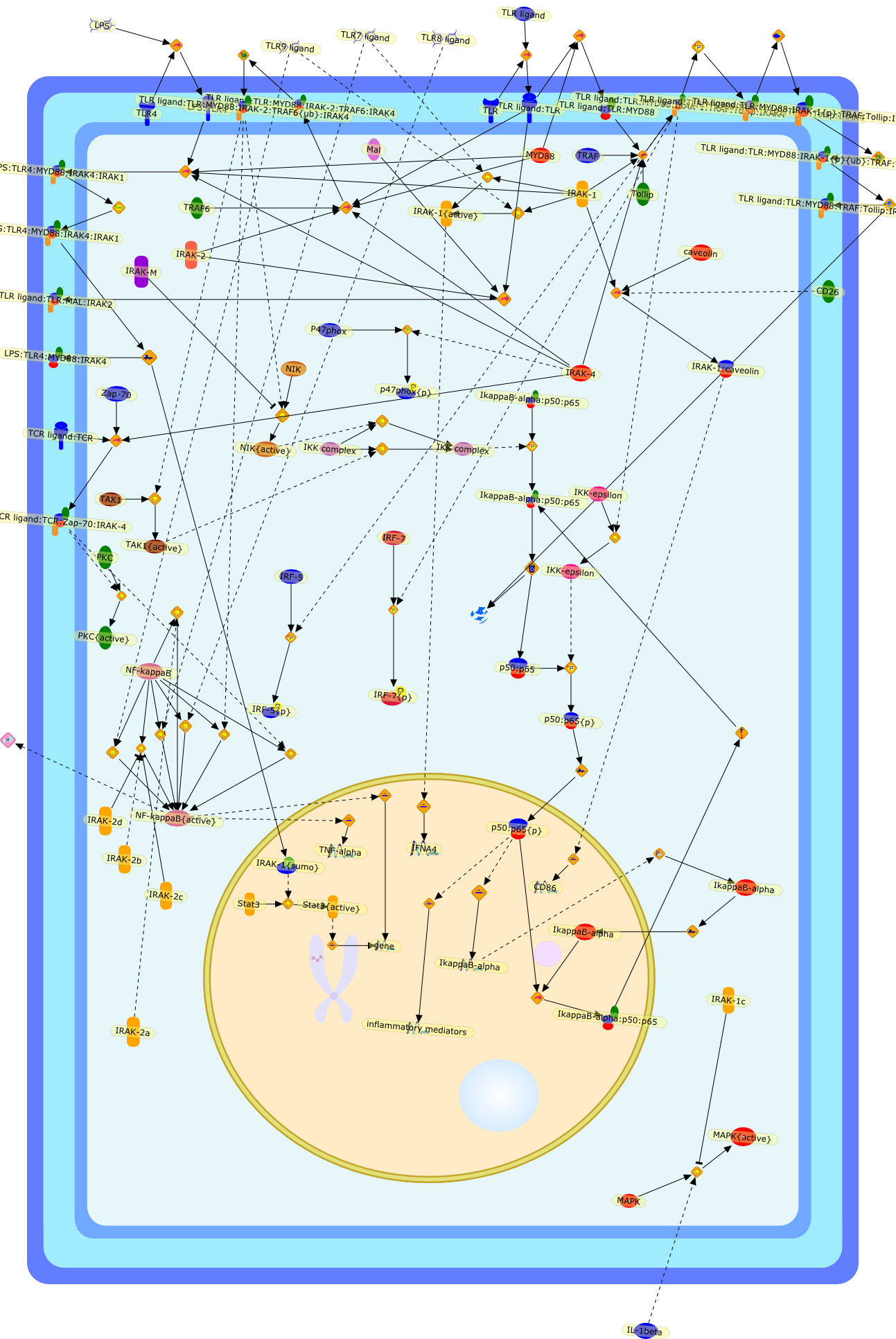
The involvement of the interleukin-1 receptor-associated kinases (IRAKs) incellular signaling networks controlling inflammation.
Affiliation
The Laboratory of Innate Immunity and Inflammation, Department of BiologicalSciences, West Campus Drive, Fralin Biotechnology Center, Virginia Tech,Blacksburg, VA 24061, USA.
Abstract
Innate immunity and inflammation plays a key role in host defense and woundhealing. However, Excessive or altered inflammatory processes can contribute tosevere and diverse human diseases including cardiovascular disease, diabetes andcancer. The interleukin-1 receptor-associated kinases (IRAKs) are criticallyinvolved in the regulation of intracellular signaling networks controllinginflammation. Collective studies indicate that IRAKs are present in many celltypes, and can mediate signals from various cell receptors includingtoll-like-receptors (TLRs). Consequently, diverse downstream signaling processescan be elicited following the activation of various IRAKs. Given the criticaland complex roles IRAK proteins play, it is not surprising that geneticvariations in human IRAK genes have been found to be linked with various humaninflammatory diseases. This review intends to summarize the recent advancesregarding the regulations of various IRAK proteins and their cellular functionsin mediating inflammatory signaling processes.
PMID
18249132
|