| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Macrophage differentiation and function in health and disease.
Affiliation
Division of Cellular and Molecular Pathology, Department of Cellular Function,Niigata University Graduate School of Medical and Dental Sciences, Niigata,Japan. mnaito@med.niigata-u.ac.jp
Abstract
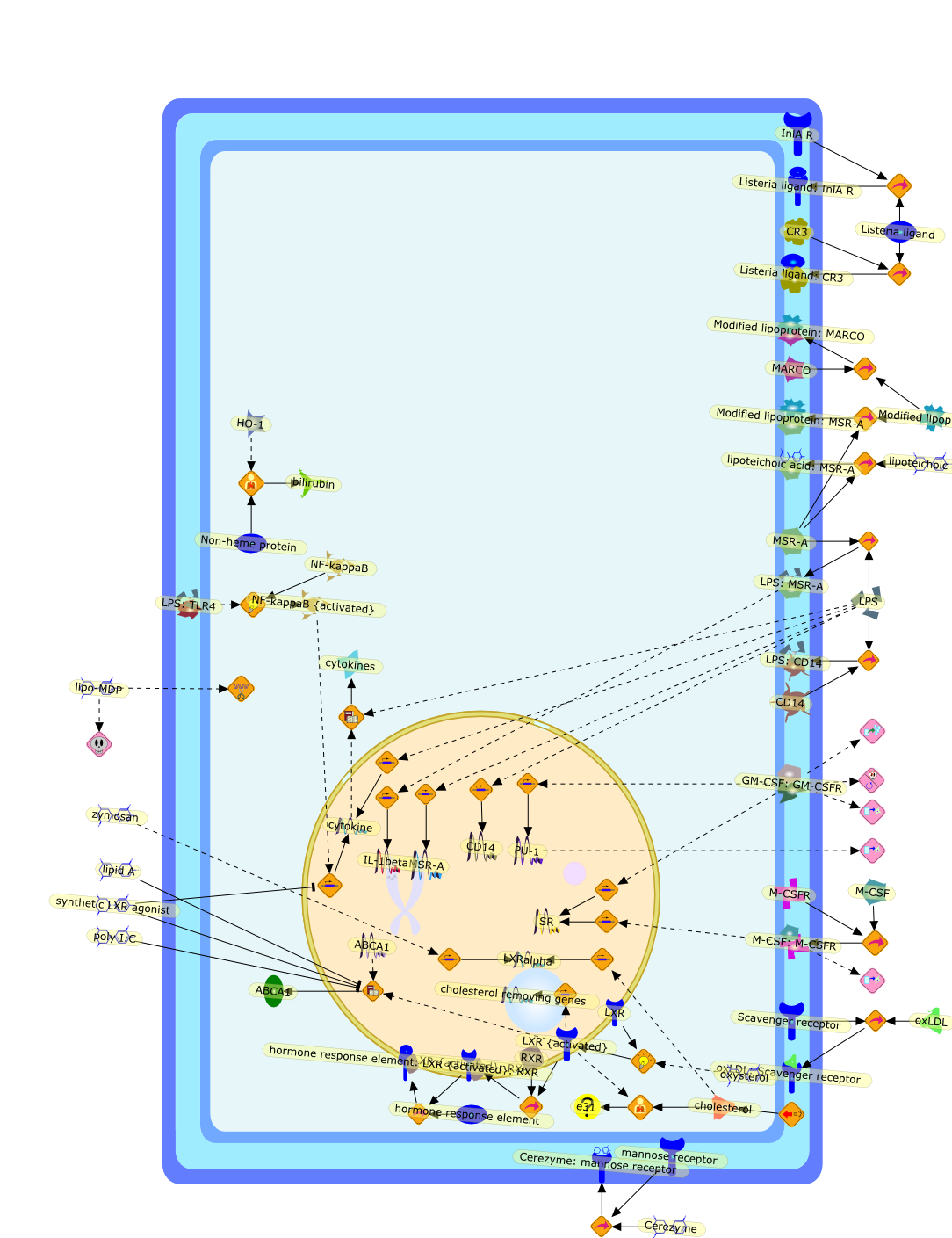
Macrophages exist in almost all animals. In some invertebrates, mesenchymalcells, endothelial cells or fibroblast-like cells can transform intomacrophages. In vertebrates, primitive macrophages first develop in yolk sachematopoiesis and differentiate into fetal macrophages. Monocytes aredifferentiated from hematopoietic stem cells in the late stage of fetalhematopoietic organs and bone marrow. Macrophages serve as an effector inmetabolism and host defense. Depletion of macrophages severely reduced bilirubinproduction and host resistance to infection. Macrophage scavenger receptors areinvolved in host defense. Macrophage growth factors are critical for macrophagedifferentiation and function. In macrophage colony-stimulating factor(M-CSF)-deficient osteopetrotic mice, monocytes, tissue macrophages andosteoclasts are deficient. Granulocyte-macrophage colony-stimulating factor(GM-CSF)-deficient mice develop alveolar proteinosis due to impaired surfactantcatabolism by alveolar macrophages. Accumulation of glucocerebroside inmacrophages due to the deficiency of glucocerebrosidase in lysosomes producesGaucher cells. Macrophages in the arterial wall incorporate chemically modifiedlow-density lipoprotein (LDL) and transform into foam cells. Binding oxidizedLDL to liver X receptor alpha (LXRalpha) upregulates the expression of itstarget genes, which act as cholesterol removers from macrophages. Inflammatorysignals downregulate the expression of LXRalpha and enhance lipid accumulation.Thus, macrophages play a pivotal role in metabolism and host defense.
PMID
18251777
|