| Original Literature | Model OverView |
|---|---|
|
Publication
Title
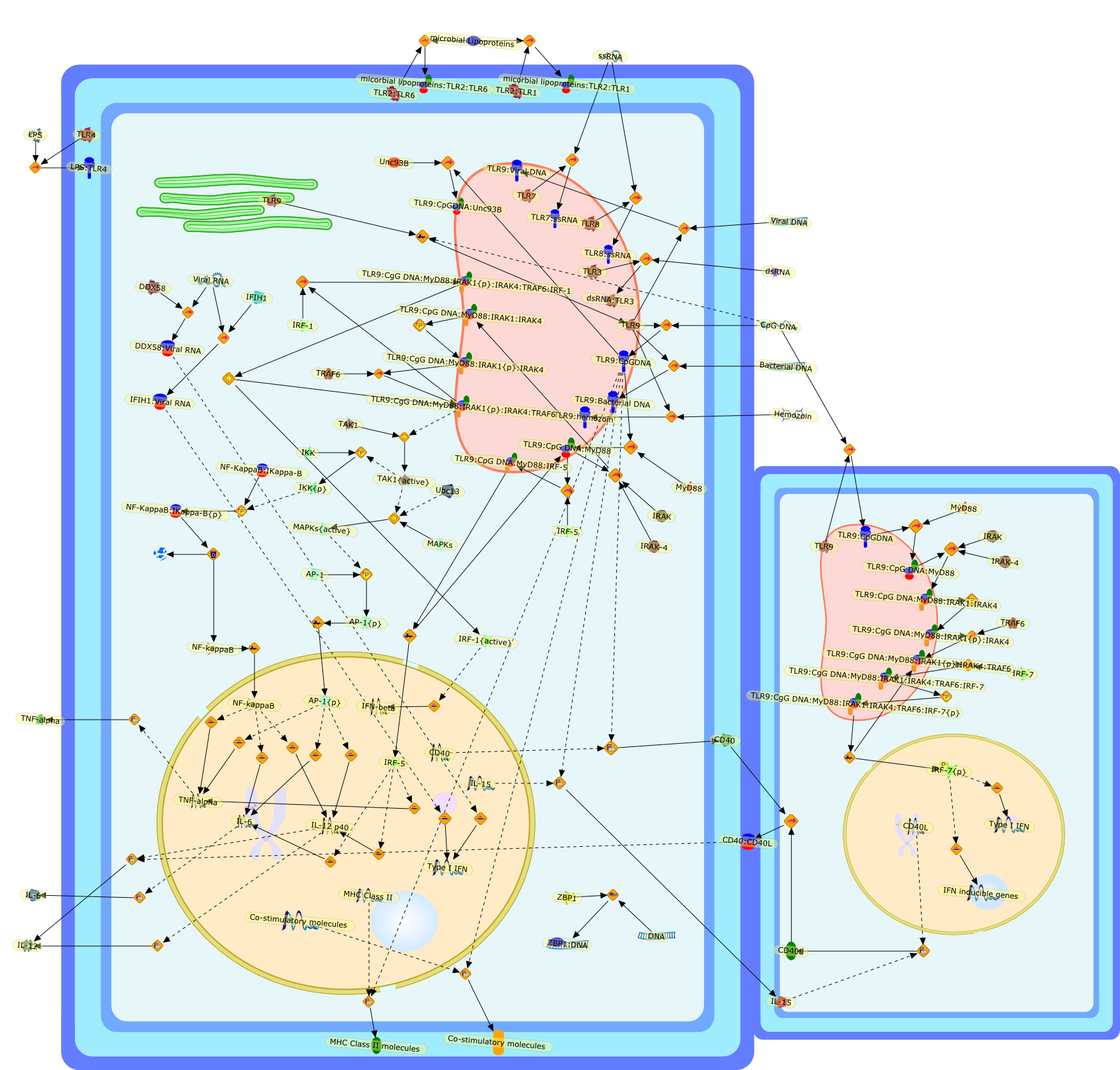
TLR9 as a key receptor for the recognition of DNA.
Affiliation
Laboratory of Host Defense, WPI Immunology Frontier Research Center, 3-1Yamada-oka, Suita, Osaka 565-0871, Japan.
Abstract
Unmethylated DNA with CpG-motifs is recognized by Toll-like receptor 9 (TLR9)and pleiotropic immune responses are elicited. Macrophages and conventionaldendritic cells (cDCs) produce proinflammatory cytokines to B/K-type CpG-DNA,whereas plasmacytoid DCs induce type I interferons to A/D-type CpG-DNA and DNAviruses. The TLR9 mediated signaling pathway is not only responsible foractivation of innate immune cells, but also for mounting acquired responses.Thus, it has been attempted to exploit TLR9 ligands as a vaccine adjuvant foranti-cancer immunotherapy. Further, TLR9 mediated signaling is implicated in thepathogenesis of autoimmune diseases such as systemic lupus erythematosus.Nevertheless, recent studies revealed that double-stranded DNA can be recognizedby intracellular receptor(s) in a TLR9-independent manner. This review willfocus on the roles of TLR9 in immune responses, and its signaling pathways.
PMID
18262306
|