| Original Literature | Model OverView |
|---|---|
|
Publication
Title
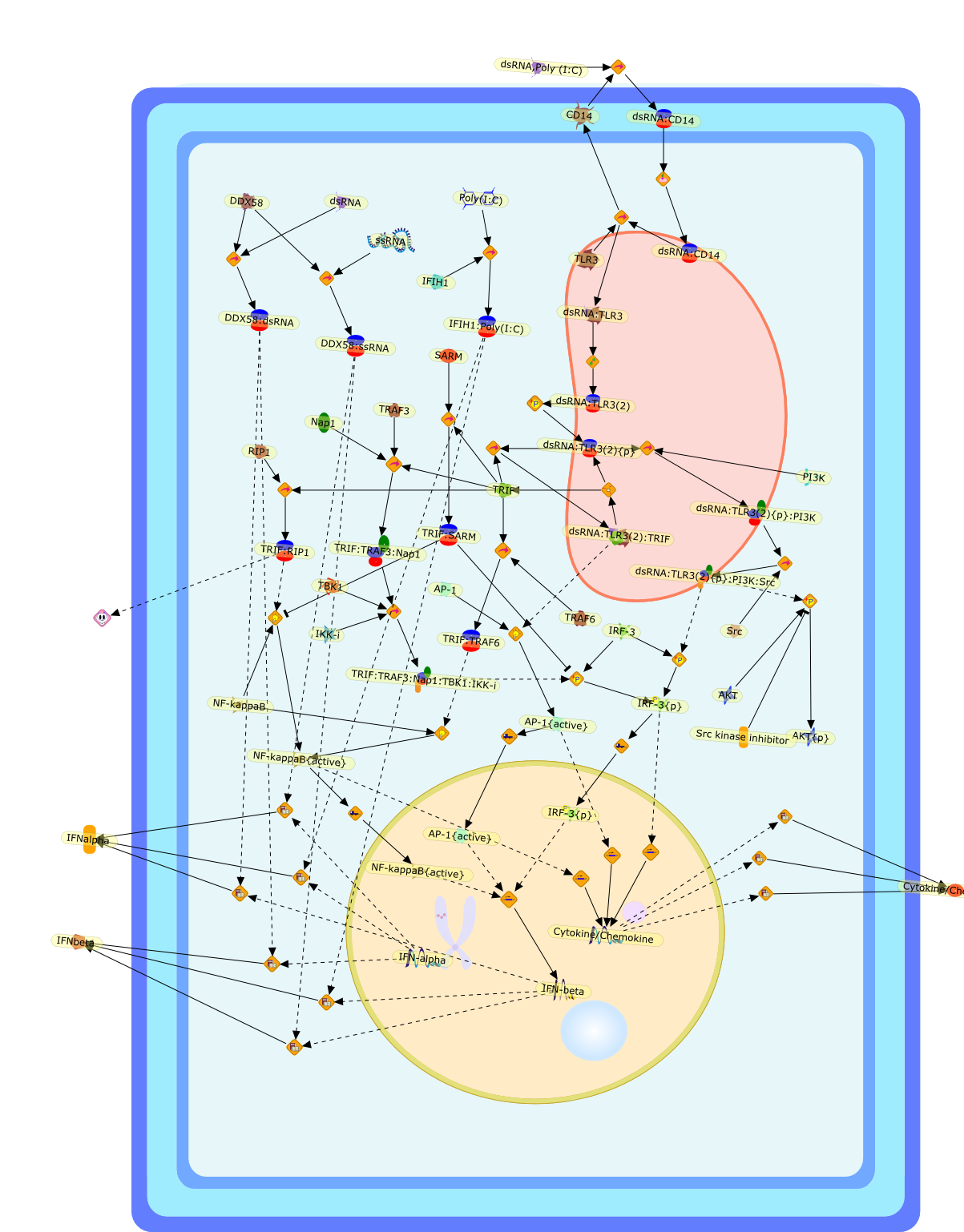
TLR3: interferon induction by double-stranded RNA including poly(I:C).
Affiliation
Department of Microbiology and Immunology, Hokkaido University Graduate Schoolof Medicine, Kita 15, Nishi 7, Kita-ku, Sapporo 060-8638, Japan.matumoto@pop.med.hokudai.ac.jp
Abstract
Toll-like receptor 3 (TLR3) recognizes viral double-stranded RNA and itssynthetic analog polyriboinosinic:polyribocytidylic acid (poly(I:C)) and inducestype I interferon (IFN), inflammatory cytokine/chemokine production anddendritic cell (DC) maturation via the adaptor protein TICAM-1 (also calledTRIF). TLR3 is expressed both intracellularly and on the cell surface offibroblasts and epithelial cells, but is localized to the endosomal compartmentof myeloid DCs. Several studies in TLR3-deficient mice demonstrate that TLR3participates in the generation of protective immunity against some viralinfections. Involvement of TLR3-TICAM-1 in activation of NK cells and CTLs bymyeloid DCs suggests that TLR3 serves as an inducer of cellular immunity sensingviral infection rather than a simple IFN inducer. In this review, we summarizethe current knowledge on TLR3 and discuss its possible role in innate andadaptive immunity.
PMID
18262679
|