| Original Literature | Model OverView |
|---|---|
|
Publication
Title
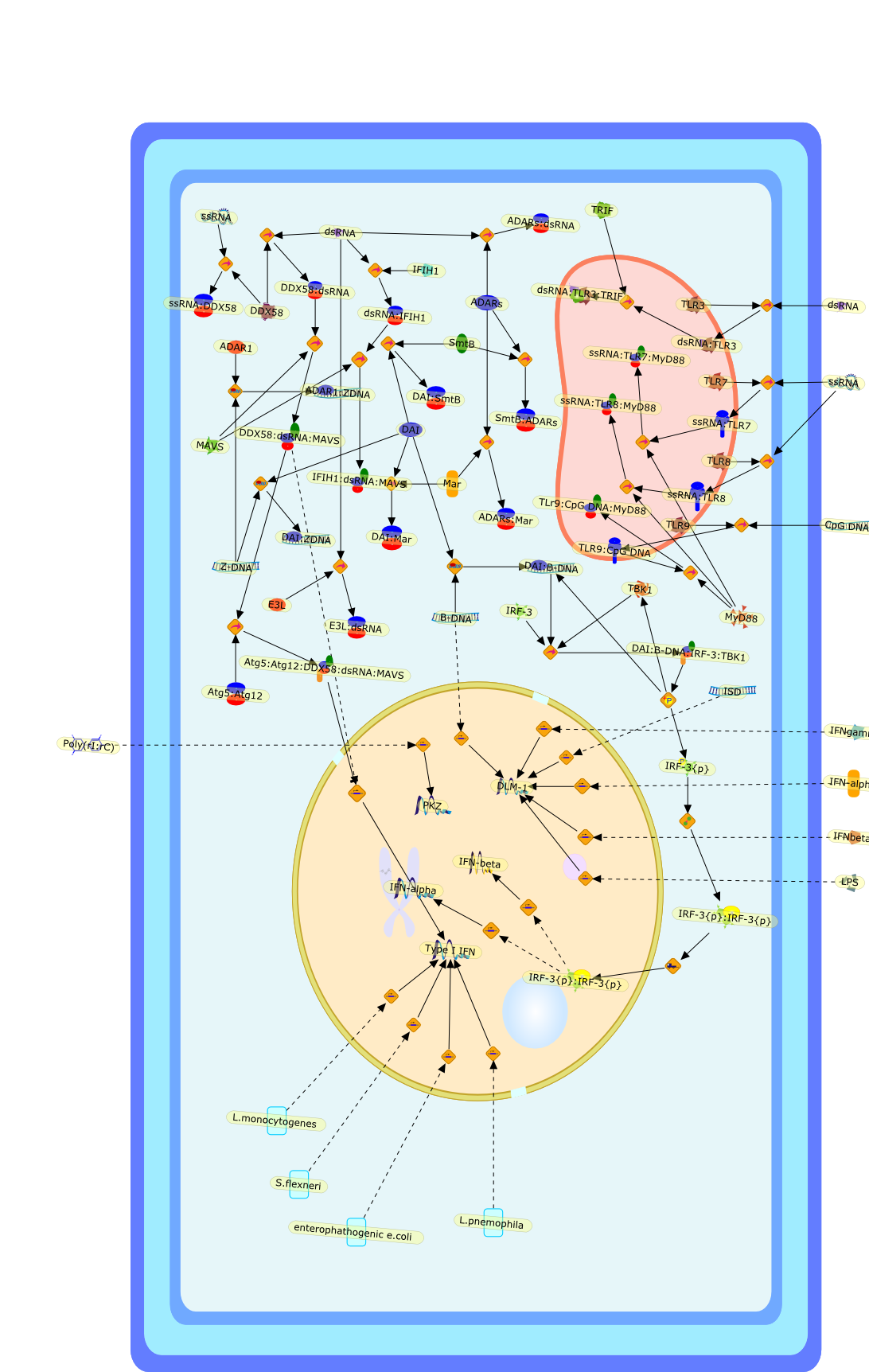
Cytosolic DNA recognition for triggering innate immune responses.
Affiliation
Division of Signaling in Cancer and Immunology, Institute for Genetic Medicine,Hokkaido University, Sapporo, Japan. takaoka@igm.hokudai.ac.jp
Abstract
The detection of microbial components by pattern recognition receptors (PRRs)and the subsequent triggering of innate immune responses constitute the firstline of defense against infections. Recently, much attention has been focused oncytosolic nucleic acid receptors; the activation of these receptors commonlyevokes a robust innate immune response, the hallmark of which is the inductionof type I interferon (IFN) genes. In addition to receptors for RNA, receptorsthat detect DNA exposed in the cytosol and activate innate immune responses havelong been thought to exist. Recently, DAI (DLM-1/ZBP1) has been identified as acandidate cytosolic DNA sensor. Cytosolic signaling by DNA-activated DAI(DLM-1/ZBP1) signaling results in activation of the two pathways of genetranscription critical to innate immune responses, the IRF and NF-kappaBpathways. In this review, we summarize our current view of activation mechanismand immunological roles of DAI (DLM-1/ZBP1) and related molecules. In addition,we also discuss the issue of self vs. non-self DNA recognition by DAI(DLM-1/ZBP1) and other DNA sensors in terms of the possible involvement inautoimmune abnormalities.
PMID
18280611
|