| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Nucleic acid-sensing Toll-like receptors: beyond ligand search.
Affiliation
Division of Infectious Genetics, Department of Microbiology and Immunology, TheInstitute of Medical Science, University of Tokyo, Japan.kmiyake@ims.u-tokyo.ac.jp
Abstract
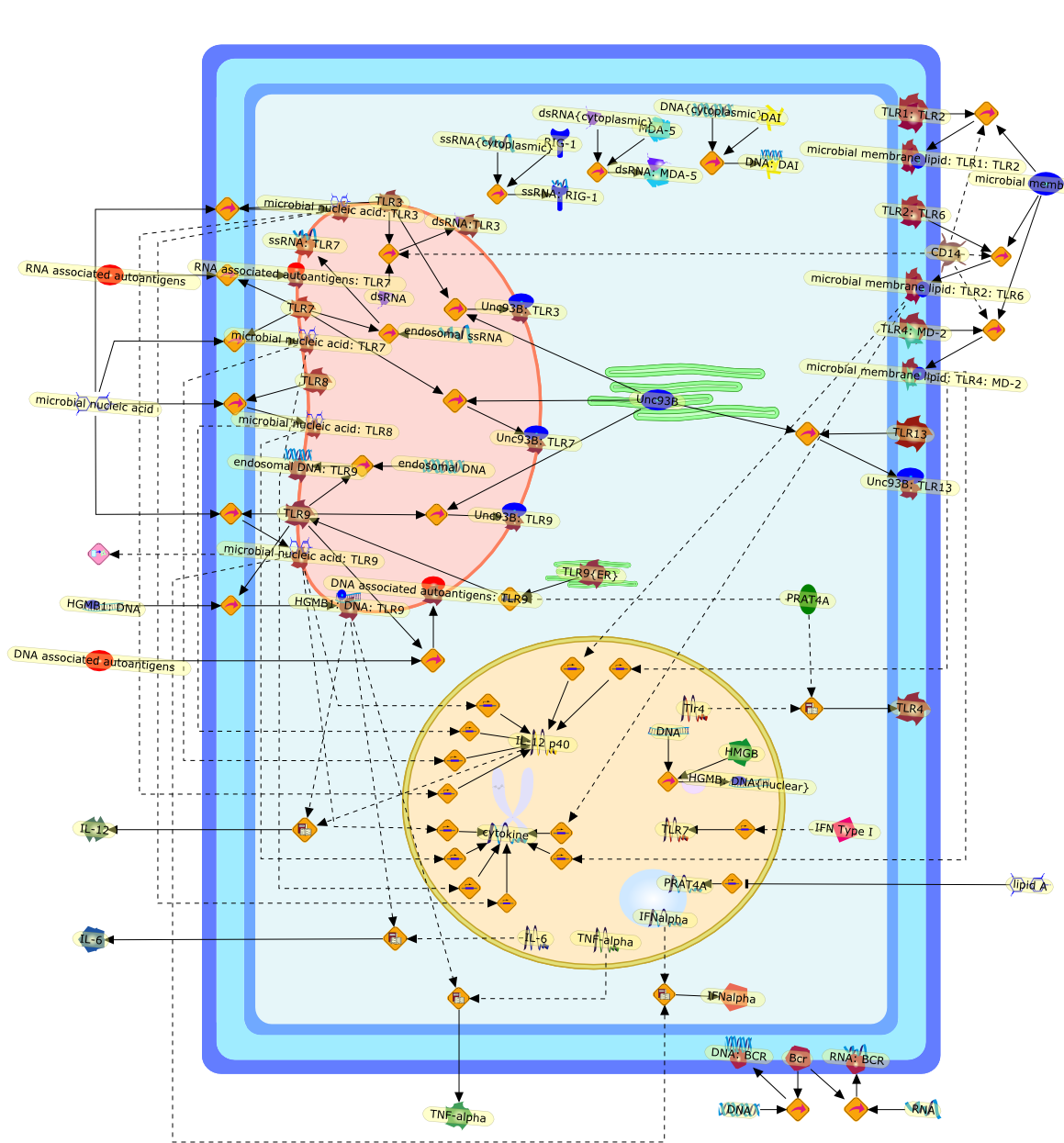
Nucleic acids from microbes were known to be immunogenic for a long time, buttheir sensors have been only recently identified. Two types of sensors,Toll-like receptors (TLR) and the RIG-I-like receptors (RLR), principallyrespond to nucleic acid. Discovery of these sensors have opened basic researchas to how our immune system handles microbial nucleic acids. Recent resultsdemonstrate that functional interaction between pathogen sensors is criticallyinvolved in the extent and type of immune responses. Moreover, responsiveness ofnucleic acid-sensing TLRs is profoundly influenced by molecules associated withTLRs or with nucleic acids. Our understanding of mechanisms by which TLRresponses were controlled would contribute to development of drugs such asimmunostimulatory nucleic acids or small chemicals antagonizing TLRs.
PMID
18321608
|