| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Mechanisms for the anti-inflammatory effects of adiponectin in macrophages.
Affiliation
Department of Pathobiology, Cleveland Clinic, Cleveland, OH 44195, USA.
Abstract
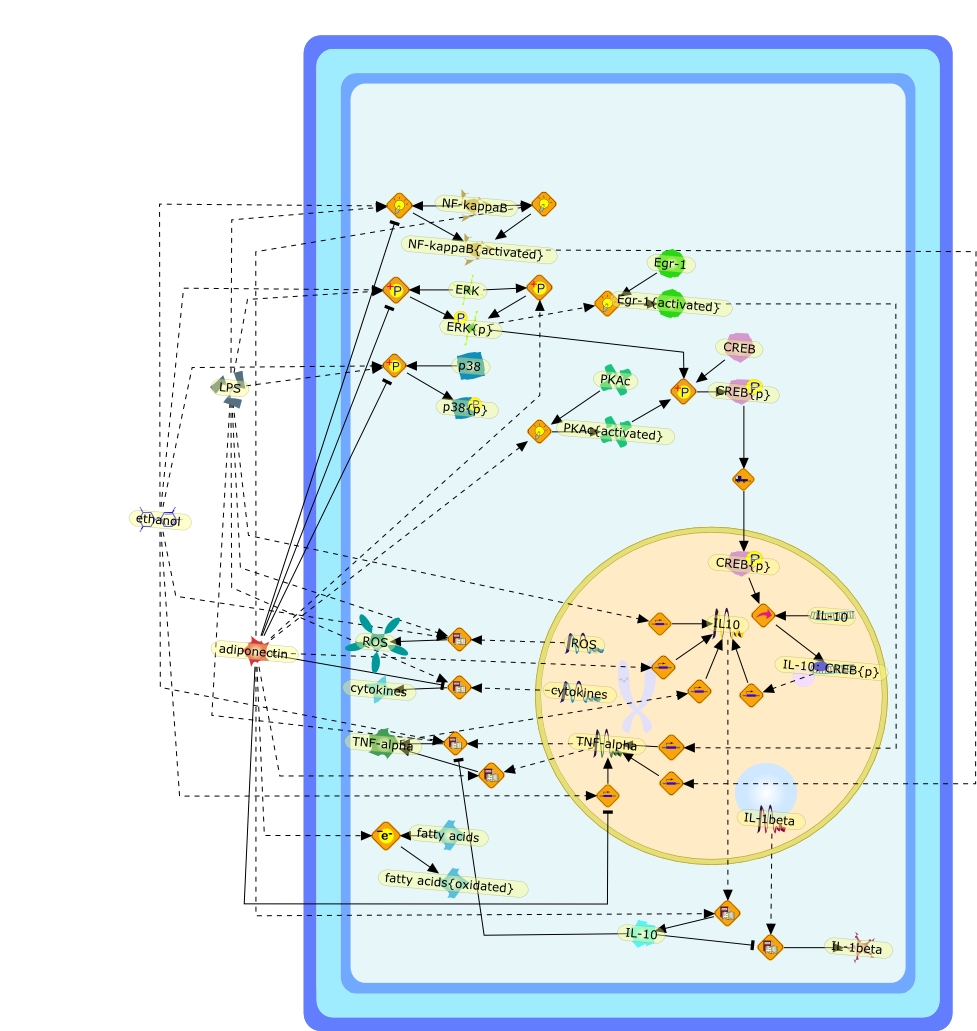
Adiponectin is an adipokine with potent anti-inflammatory properties. Thedevelopment of alcoholic liver disease is thought to involve increasedpro-inflammatory activity, mediated in part by the activation of hepaticmacrophages (Kupffer cells). Chronic ethanol feeding sensitizes hepaticmacrophages to activation by lipopolysaccharide (LPS), leading to increasedproduction of reactive oxygen species and tumor necrosis factor-alpha(TNF-alpha). Adiponectin can normalize Toll-like receptor-4 (TLR-4) mediatedsignaling in hepatic macrophages after ethanol feeding, likely contributing tothe hepatoprotective effect of adiponectin in the progression of alcoholic liverdisease. However, the mechanisms by which adiponectin suppress TLR-4 mediatedresponses are not well understood. Using the macrophage-like cell line,RAW264.7, we have investigated the molecular mechanisms by which adiponectinsuppresses LPS-stimulated TNF-alpha production. Globular adiponectin(gAcrp)-mediated desensitization of LPS-stimulated responses in RAW264.7macrophages was dependent on the production of the anti-inflammatory cytokineinterleukin (IL)-10. gAcrp initially increased TNF-alpha expression in RAW264.7macrophages; this TNF-alpha then contributed to increased expression of IL-10.This initial gAcrp-mediated increase in TNF-alpha production by macrophages wasmediated via activation of ERK1/2-->Egr-1 and nuclear factor(NF)-kappaB-dependent mechanisms. gAcrp-stimulated IL-10 expression was alsodependent on the phosphorylation of cAMP response element-binding protein andthe cAMP response element in the IL-10 promoter. In summary, these studiesreveal a complex, integrated response of macrophages to gAcrp. gAcrp initiallyactivated signaling pathways considered to be pro-inflammatory, with asubsequent increase in the expression of the potent, anti-inflammatory cytokine,IL-10. Increased IL-10 expression was ultimately required for the suppression ofTLR4-mediated signaling by gAcrp.
PMID
18336664
|