| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Lipopolysaccharide sensing an important factor in the innate immune response toGram-negative bacterial infections: benefits and hazards of LPShypersensitivity.
Affiliation
Max-Planck-Institute for Immunobiology, Stubeweg 51, D-79108 Freiburg, Germany.freudenberg@immunobio.mpg.de
Abstract
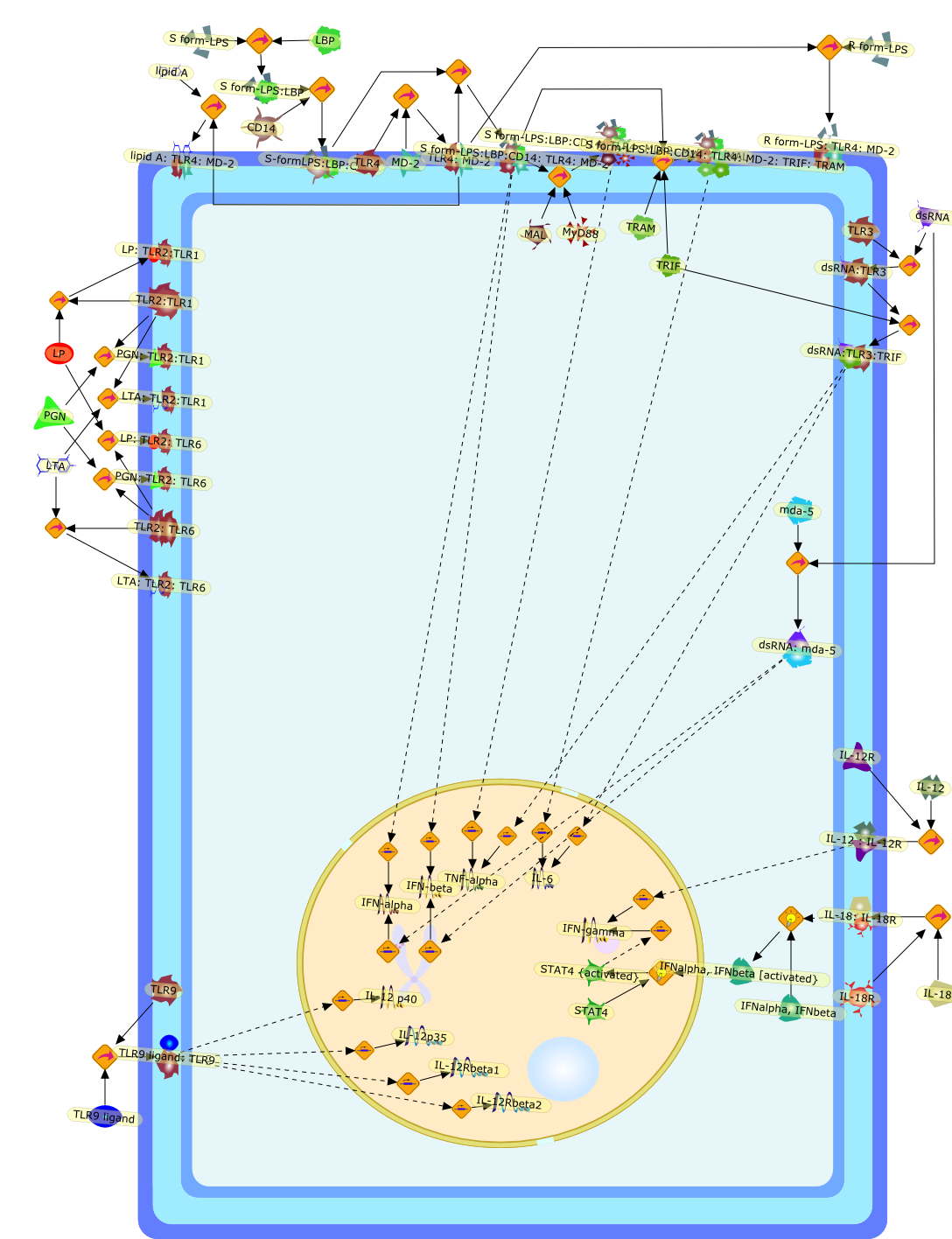
In this review, we summarize our investigations concerning the differentialimportance of CD14 and LBP in toll-like receptor 4 (TLR4)/myeloiddifferentiation protein-2 (MD-2)-mediated signaling by smooth and rough-formlipopolysaccharide (LPS) chemotypes and include the results obtained in studieswith murine and human TLR4-transgenic mice. Furthermore, we present more recentdata on the mechanisms involved in the induction of LPS hypersensitivity bybacterial and viral infections and on the reactivity of the hypersensitive hostto non-LPS microbial ligands and endogenous mediators. Finally, the effects ofpre-existing hypersensitivity on the course and outcome of a super-infectionwith Salmonella typhimurium or Listeria monocytogenes are summarized.
PMID
18406367
|