| Original Literature | Model OverView |
|---|---|
|
Publication
Title
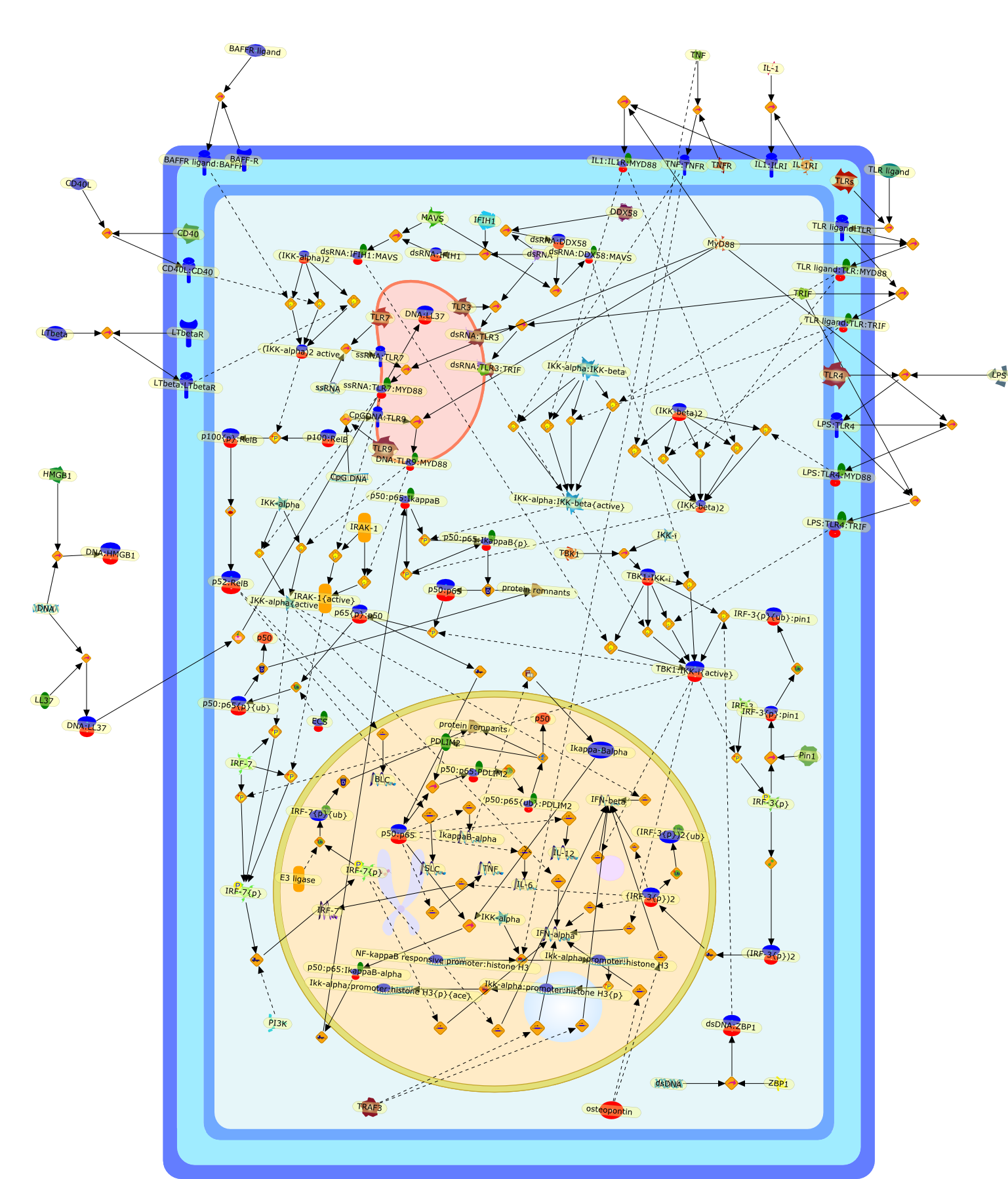
Turning NF-kappaB and IRFs on and off in DC.
Affiliation
Laboratory for Host Defense, RIKEN Research Center for Allergy and Immunology,Yokohama city, Kanagawa 230-0045, Japan. tkaisho@rcai.riken.jp
Abstract
Dendritic cells (DCs) produce an array of cytokines after detecting variousimmune adjuvants through pattern recognition receptors (PRRs). PRR signalingleads to activation of transcription factors such as NF-kappaB or interferonregulatory factors (IRFs) but after activation must be attenuated to avoidimmunopathology and to maintain tissue homeostasis. IkappaB kinase familymembers, originally identified as classical NF-kappaB activators, are now foundto be broadly and crucially involved in PRR signaling in a member-specificmanner. Furthermore, a new mechanism for NF-kappaB downregulation is emergingthat involves the degradation of active NF-kappaB by the nuclearubiquitin-proteasome system. Here we review new aspects of NF-kappaB and IRFregulation chiefly in DCs.
PMID
18534908
|