| Original Literature | Model OverView |
|---|---|
|
Publication
Title
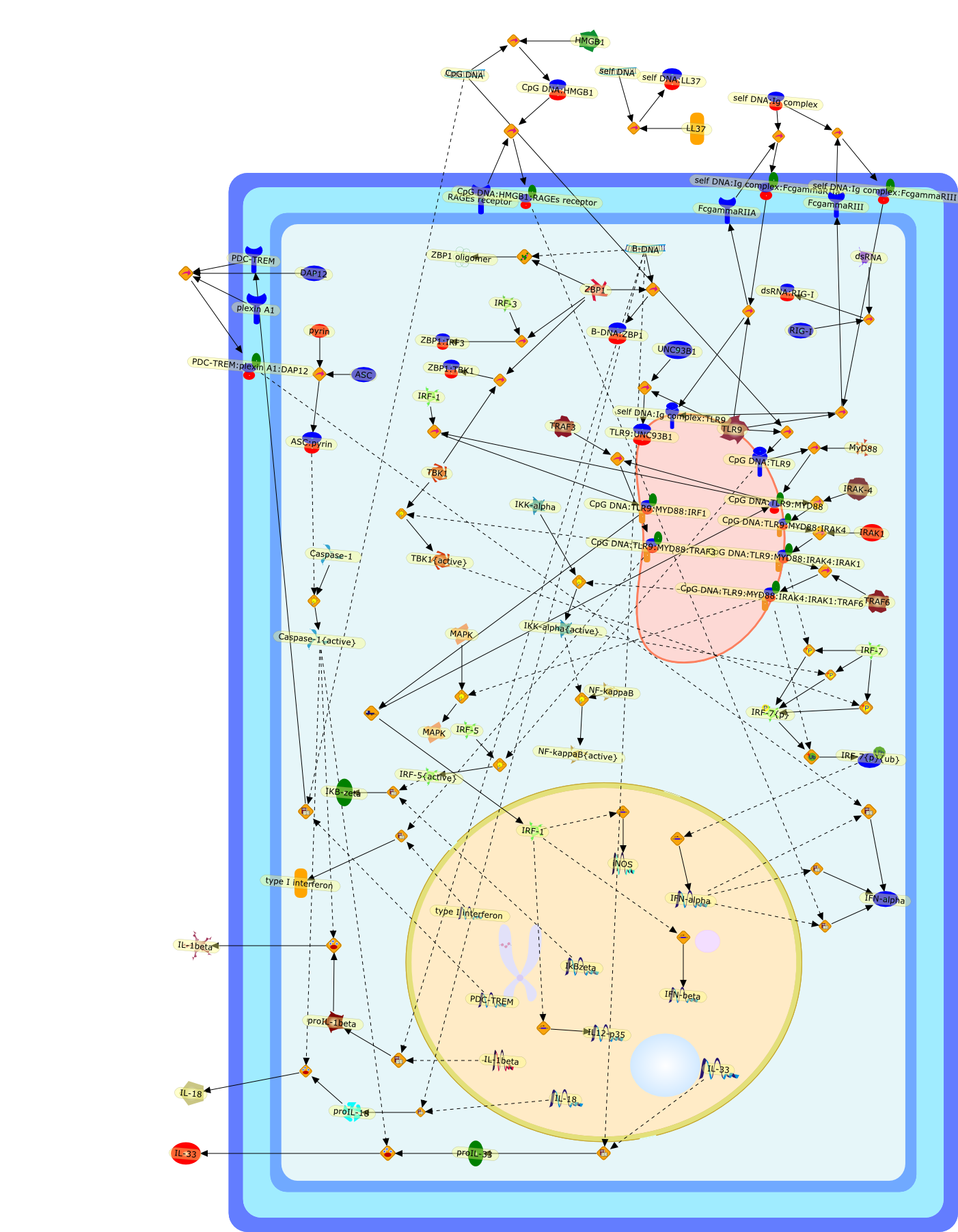
Intracellular DNA sensors in immunity.
Affiliation
Department of Molecular Biodefense Research, Yokohama City University GraduateSchool of Medicine, 3-9 Fukuura, Kanazawaku, Yokohama, Japan.takesita@yokohama-cu.ac.jp
Abstract
Mammalian innate immunity possesses a distinct system to recognize aberrant DNAinside the cell. One class of DNA sensors is the Toll-like receptor 9, which isexpressed in the specialized immune cells, binds to single-stranded DNA in theendosome to transmit cellular signaling through myeloid differentiation primaryresponse protein 88 (MyD88). Another class of DNA sensors exists in thecytoplasm of most type of cells in the tissue, detecting double-stranded DNA tosignal through TANK-binding kinase-1 (TBK1)-mediated type-I interferonproduction and apoptosis-associated speck-like protein containing a CARD(ASC)-mediated IL-1beta secretion. Since DNA sensors have potential to recognizeaberrant DNA of both self and nonself origin, their physiological roles inmicrobial infection, tissue damage, autoimmune diseases, and DNA-basedtherapeutic applications are being intensively investigated.
PMID
18573338
|