| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Signalling adaptors used by Toll-like receptors: an update.
Affiliation
School of Biochemistry and Immunology, Trinity College, Dublin, Ireland.kennyef@tcd.ie
Abstract
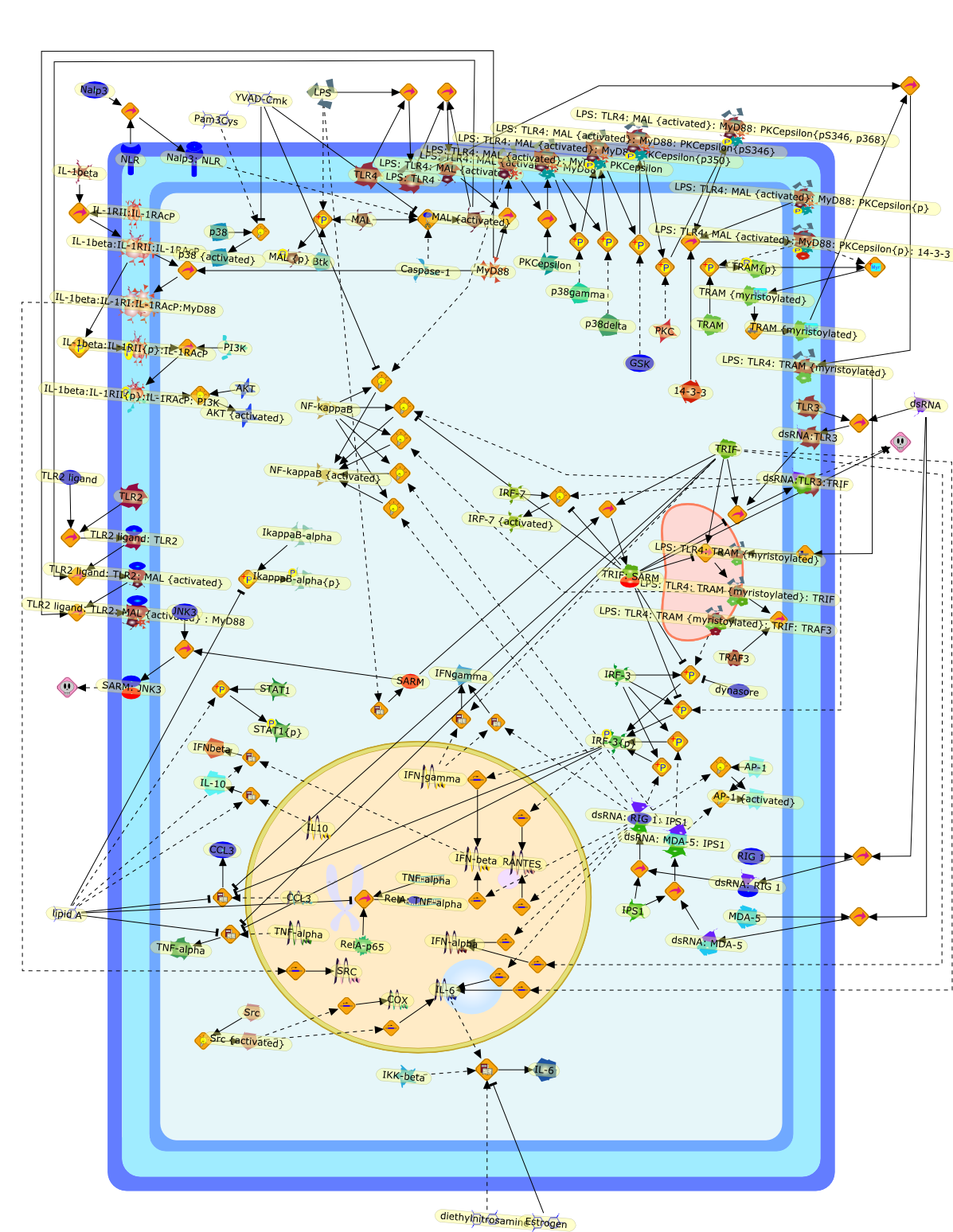
Research into the five Toll/IL1 receptor (TIR) adaptor proteins involved ininnate immunity continues to advance. Here we outline some of the more recentfindings. MyD88 has a key role in signalling by the IL1 receptor complex andTLRs. However, a MyD88-independent pathway of IL1beta signalling in neurons hasbeen described which involves the protein kinase Akt, and which has ananti-apoptotic effect. This pathway may also be important for the mechanismwhereby Alum exerts its adjuvant effect since this depends on IL1beta but isMyD88-independent. MyD88 is also involved in tumourigenesis in models ofhepatocarcinoma and familial associated polyposis (FAP); negative regulation ofTLR3 signalling and in PKCepsilon activation. The adaptor Mal is regulated byphosphorylation and caspase-1 cleavage. A variant form of Mal in humans termedS180L confers protection in multiple infectious diseases. TRAM is controlled bymyristoylation and phosphorylation and the localisation of TRAM with TLR4 toendosomes is required for activation of IRF3 and induction of IFNbeta. FinallySARM has been shown to regulate TRIF and also appears to be involved in neuronalinjury mediated by oxidative stress in mouse neurons. These advances confirm theimportance for the TIR domain-containing adapters in host defence andinflammation.
PMID
18706831
|