| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Acetylation of MKP-1 and the control of inflammation.
Affiliation
Department of Immunology, St. Jude Children's Research Hospital, Memphis, TN38105, USA. hongbo.chi@stjude.org
Abstract
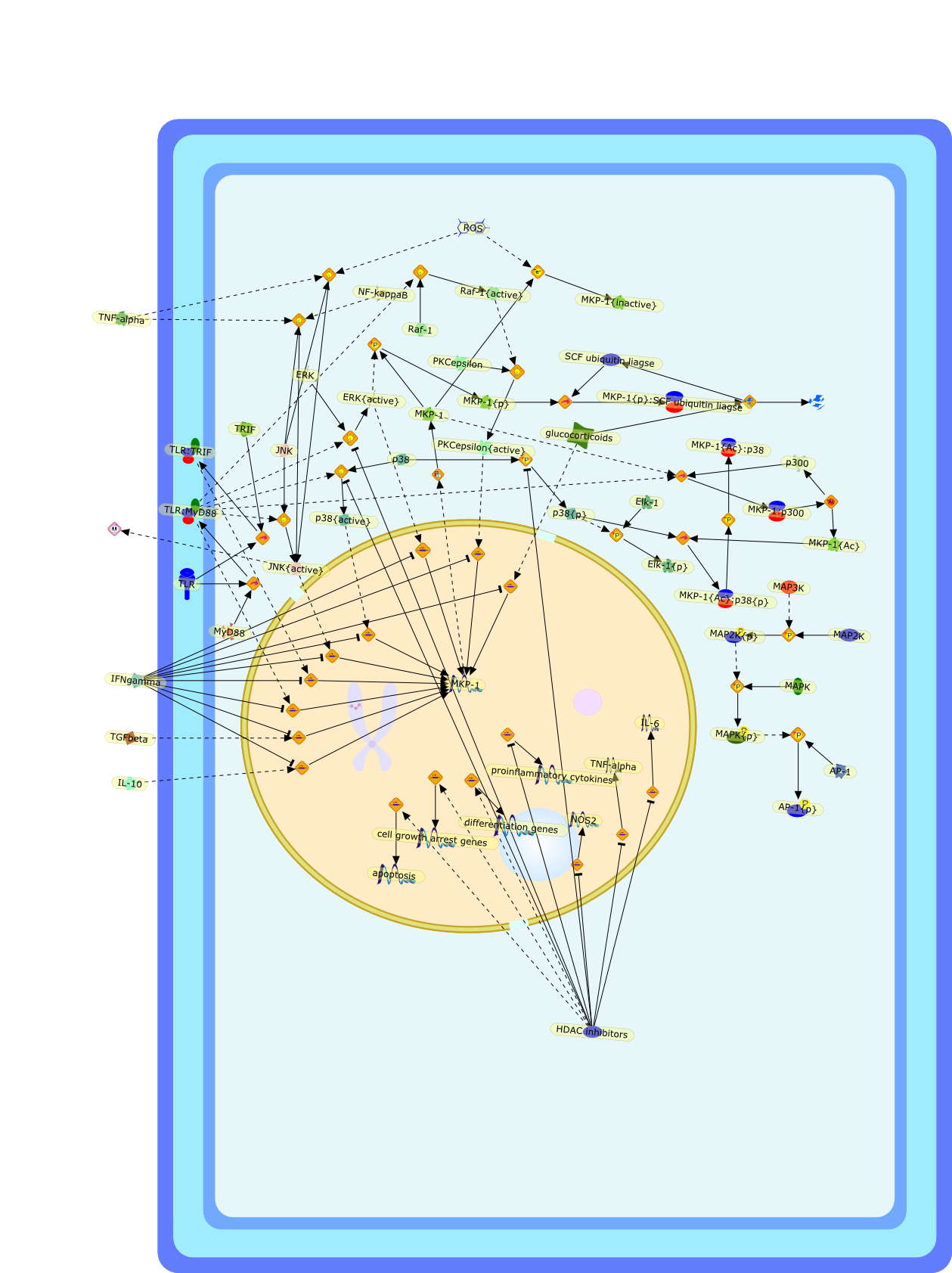
Innate immune responses mediated by Toll-like receptors (TLRs), a class ofpattern-recognition receptors, play a critical role in the defense againstmicrobial pathogens. However, excessive TLR-mediated responses result in sepsis,autoimmunity, and chronic inflammation. To prevent deleterious activation ofTLRs, cells have evolved multiple mechanisms that inhibit innate immunereactions. Stimulation of TLRs induces the expression of the gene encoding themitogen-activated protein kinase (MAPK) phosphatase-1 (MKP-1), anuclear-localized dual-specificity phosphatase that preferentiallydephosphorylates p38 MAPK and c-Jun N-terminal kinase (JNK), resulting in theattenuation of TLR-triggered production of proinflammatory cytokines. MKP-1 isposttranslationally modified by multiple mechanisms, including phosphorylation.A study now demonstrates that MKP-1 is also acetylated on a key lysine residuefollowing stimulation of TLRs. Acetylation of MKP-1 promotes the interaction ofMKP-1 with its substrate p38 MAPK, which results in dephosphorylation of p38MAPK and the inhibition of innate immunity.
PMID
18922786
|