| Original Literature | Model OverView |
|---|---|
|
Publication
Title
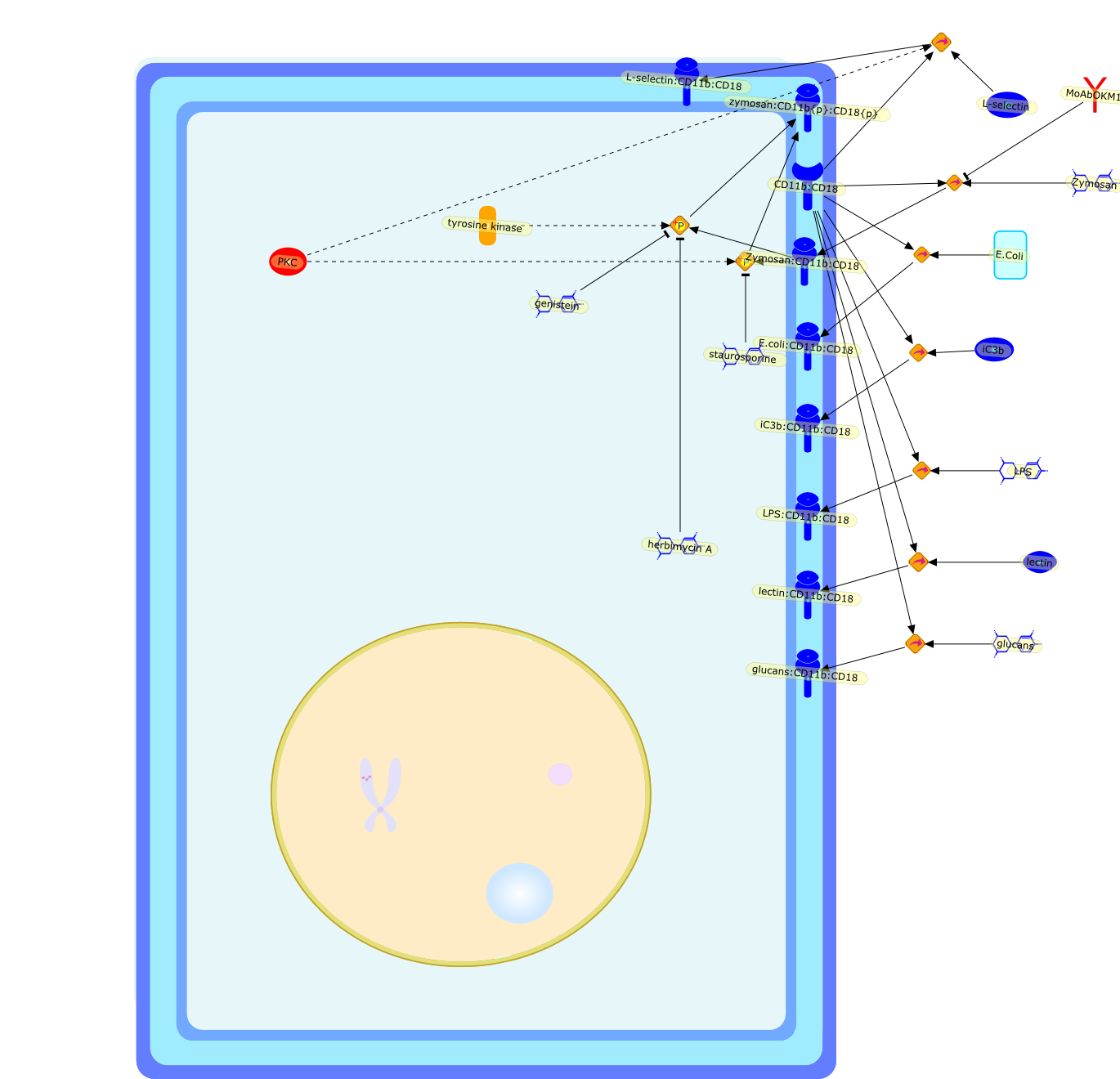
CR3 (CD11b, CD18): a phagocyte and NK cell membrane receptor with multipleligand specificities and functions.
Affiliation
Department of Microbiology and Immunology, University of Louisville, KY 40292.
Abstract
The C3 receptor CR3 is expressed on phagocytic cells, minor subsets of B and Tcells, and natural killer (NK) cells. It has important functions both as anadhesion molecule and a membrane receptor mediating recognition of diverseligands such as intercellular adhesion molecule-1 (ICAM-1) and fixed iC3b. Thereceptor is capable of undergoing an activation event that regulates both itsspecificity for various ligands and its ability to mediate phagocytosis orextracellular cytotoxicity. Certain bacteria express carbohydrates orlipopolysaccharides (LPS) that can bind to and activate CR3, allowing thereceptor to assume its activated state. Soluble beta-glucan derived from theyeast Saccharomyces cerevisiae is a particularly potent stimulator of CR3, andproduces an activated state of the receptor that permits neutrophil phagocytosisof iC3b-coated erythrocytes or NK, cell cytotoxicity of iC3b-coated tumourcells, that are normally resistant to NK cells.
PMID
8485905
|