| Original Literature | Model OverView |
|---|---|
|
Publication
Title
CSF-1 and cell cycle control in macrophages.
Affiliation
University of Melbourne, Department of Medicine, Royal Melbourne Hospital,Parkville, Victoria, Australia.
Abstract
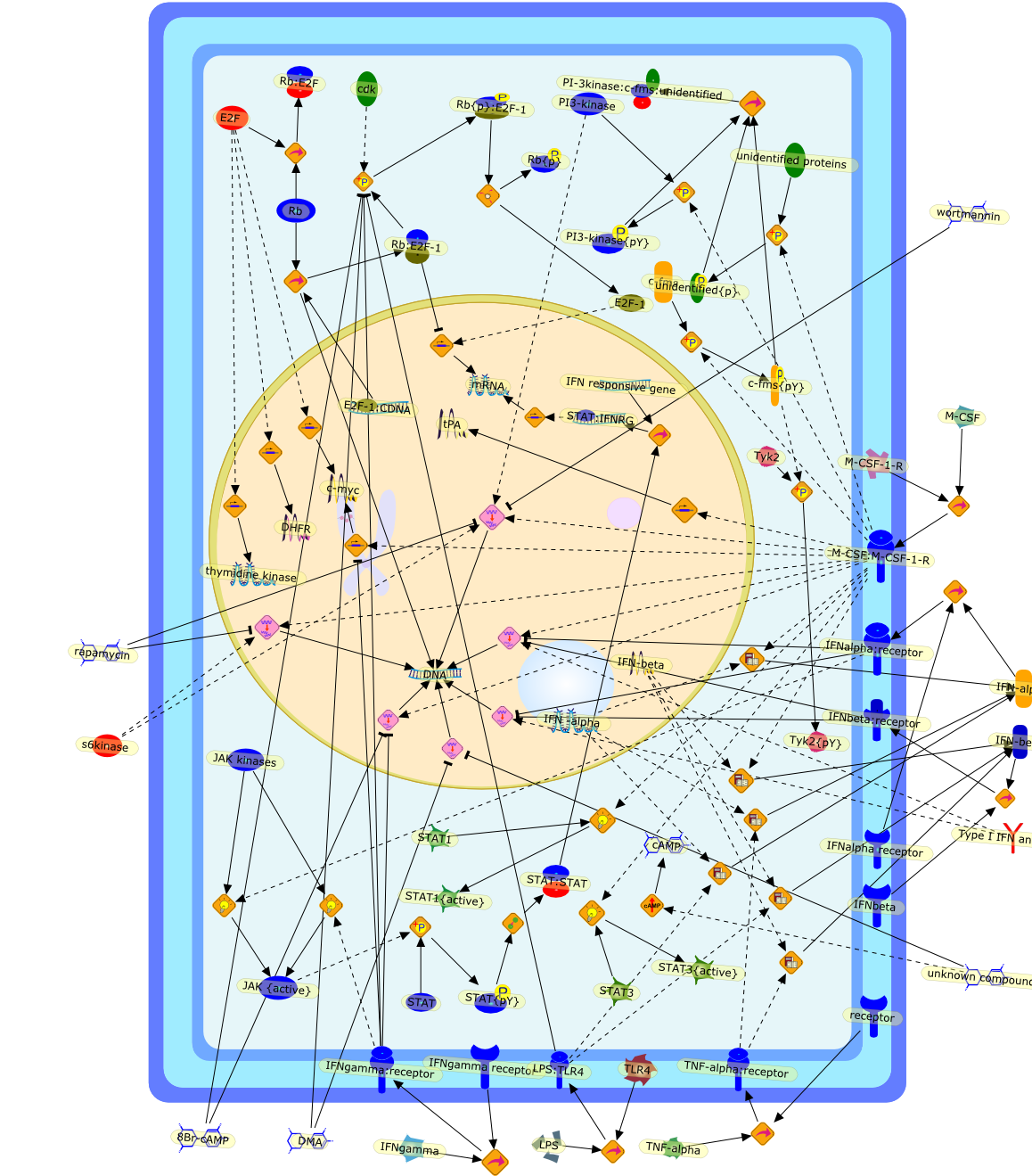
Control of cell proliferation involves a finely interwoven network of positiveand negative cell cycle regulators. Signal transduction pathways linking c-fms(CSF-1R) to cellular proliferation and differentiation are being explored. Partof the strategy is to use a series of G1 inhibitors to help pinpoint relevanttargets. Several inhibitors-8Br-cAMP, interferon gamma (IFN gamma), INFalpha/beta, lipopolysaccharide (LPS), tumor necrosis factor-alpha (TNF alpha),and dimethylamiloride-suppress CSF-1-stimulated proliferation in murine bonemarrow-derived macrophages (BMM) even when added in the mid- to late-G1 phase ofthe cell cycle. The down-modulating effects of the inhibitors on the expressionof the following cell cycle regulators have been examined: c-myc, cyclin D1 andD2, cdk4, Rb phosphorylation, E2F binding activity, ribonucleotide reductasesubunits, and PCNA. Some differences in the negative control of such regulatorswere found, for example, in the manner in which IFN gamma and cAMP down-regulatec-myc expression. Using blocking antibodies and BMM from type I IFN receptorknockout mice, it appears that one of these inhibitors, IFN alpha/beta, acts asan endogenous inhibitor in CSF-1-treated BMM and is also responsible, at leastin part, for the inhibition of cell cycle progression by LPS and TNF alpha.Another strategy has been to attempt to relate early biochemical changes inducedby CSF-1 to later changes in the G1 phase, partly by studying cycling versusnoncycling macrophages and partly by using cells expressing c-fms with tyrosinemutations in the intracytoplasmic region. CSF-1-mediated effects on thefollowing signal transduction molecules in these systems will be described:PI3-kinase, myelin basic protein kinases, Erks, and STAT transcription factors.
PMID
8981359
|